Metabolic models make remediation more manageable
(Â鶹ÒùÔºOrg.com) -- In efforts to reduce contamination at a former uranium mill tailings site, Dr. Krishna Mahadevan is developing genome-scale models to determine why certain bacteria reduce uranium better than others. The University of Toronto professor is part of a scientific team studying the Department of Energy's Integrated Field-Scale Subsurface Research Challenge site in Rifle, Colorado. He collaborates with Dr. Derek Lovley at the University of Massachusetts and Pacific Northwest National Laboratory scientists Dr. Timothy Scheibe and Dr. Philip Long.
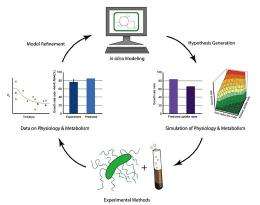
Mahadevan, an expert in the areas of systems biology, metabolic modeling, and bioprocess optimization, presented highlights of his work May 12 as part of PNNL's Frontiers in Biological Sciences Seminar Series. The seminars allow experts to share results of studies and novel ideas in biological sciences. Mahadevan presented different ways to use genome-scale metabolic models to predict outcomes in complex environmental situations such as the earth's subsurface.
Why genome-level metabolic models?
A wide diversity of unexplored metabolism is encoded in the genomes of microorganisms that have an important environmental role-such as contaminant reduction. These genomes have been the basis for the reconstruction of detailed metabolic networks and models for several microorganisms. Genome-scale metabolic modeling enables the individual reactions encoded in annotated genomes to be organized by scientists into a coherent whole, which can then be used to predict metabolic fluxes that will optimize cell function under a range of conditions that are expected in the environment and link them to "omics" data.
Mahadevan and colleagues have focused on genome-scale metabolic modeling of Geobacter, a bacterial species with important applications in bioremediation at the Rifle site and microbial fuel cells. Their research has resulted in an in-depth understanding of the species' central metabolism.
"Taking a similar modeling and experimental approach with other microorganisms that inhabit a variety of environments could speed up understanding of their physiology and ecology, and could help scientists optimize practical applications of these species," said Mahadevan.
Scalable models of biological networks expressed by different organisms will be crucial for optimizing these systems for industrial biotechnology, as well as for understanding the behavior and evolution of biological systems and their responses to multiple environmental cues. Such fundamental understanding forms the basis for extending biological design principles in more complex systems such as microbial communities, multi-cellular tissues and, in the future, to higher-level organisms.
More information:
Mahadevan R, et al. 2011. "In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modeling." Nature Reviews Microbiology 9, 39-50.
Fang Y, et al. 2010. "Direct coupling of a genome-scale microbial in silico model and a groundwater reactive transport model." Journal of Contaminant Hydrology 122(1-4):96-103.
Provided by Pacific Northwest National Laboratory