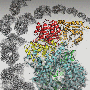Discovery of molecular machine that drives spread of flu virus
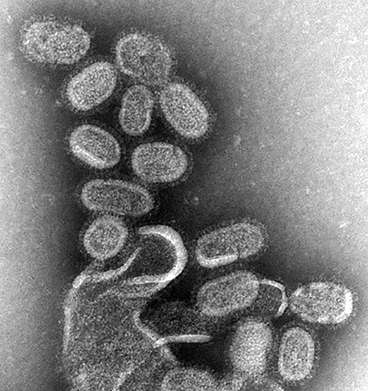
Scientists have used Diamond to uncover the structure of a key component involved in the spread of influenza C virus. Integral to viral replication, polymerases are enzymes that copy the viral genome and produce messenger RNA which is then used to make building blocks for new virus particles. Influenza C is a variety of flu, a viral infection that altogether affects approximately 3 to 5 million people every year, leading to between 250,000 to 500,000 deaths around the world. Understanding the atomic structure of this polymerase could be a major step forward, supporting the development of drugs that prevent it from functioning, effectively 'disarming' the flu virus and making it unable to spread.
Professor Ervin Fodor is part of Oxford University's Dunn School of Pathology. He observes: "The polymerase we have mapped is key to the virus's ability to replicate. If we can understand its structure, we can better understand how it works. This means we could be able to create drugs that target this polymerase to prevent the virus spreading. Understanding the structure also means we may be better placed to understand the processes by which bird flu viruses adapt to infect humans and cause viral outbreaks."
The collaboration of scientists from Oxford University used Diamond X-ray capabilities to unpick the structure of the influenza C virus polymerase. The enzyme is made up of three chemically different chains of amino acids. The key breakthrough in this work was to produce and crystallise the entire polymerase structure in a conformation not previously known. This research is funded by the Medical Research Council and the Wellcome Trust.
Dr Megan Dowie, programme manager for structural studies at the MRC, said: "This study reveals the detailed structure of an important molecular machine within a flu virus. By visualising it in an inactive state, the researchers have identified a potential new treatment target, as keeping the polymerase in this state would affect the virus's ability to replicate. The Diamond synchrotron is a powerful tool for researchers to gain valuable molecular insights into a range of diseases."
Prof Jonathan Grimes is a Diamond Fellow and Associate Professor of Structural Biology at the University of Oxford. He uses Diamond to study a variety of viruses, and is particularly interested in how they replicate. Prof Grimes adds: "Polymerases are just one component of the complex replication process. Research facilities at Diamond enable us to map and visualise these tiny biological processes at the atomic scale. With this valuable information, pharmaceutical research - whether drug design or vaccine development - can be better targeted. Knowledge like this is one of the first steps towards the creation of novel medical approaches and treatments"
Importantly, this finding could also impact on the development of drugs to treat similar viruses.
Prof Grimes concludes: "Whilst viruses differ in terms of their structure, effect, and components, just like other species they can be grouped into families based on shared characteristics. Understanding how the polymerase in influenza C virus is structured and functions could provide insight into polymerases of other viruses in the same family, including the more aggressive and dangerous influenza A and B virus types. From this knowledge, we will also study the polymerases for rabies and Ebola in an attempt to determine their exact structure, so that new avenues for the treatment of these diseases can be opened up in a similar way."
The discovery of the influenza C virus polymerase structure is an important step forward for both treating flu and for understanding these molecular machines and how they can be targeted to halt the spread of viruses more generally.
More details about the structure solved by scientists
Influenza C polymerase is a key enzyme also known as FluPol. This complex structure is made up of three proteins that are bound to each of the RNA genome segments of the virus. These RNA genome segments contain the genetic information of the virus. When a virus enters a cell, FluPol – together with the bound RNA – is released into the cell nucleus where it creates new copies of the genomic RNA to be packaged into new virus particles. In a different process, FluPol also steals a key part of the cells' messenger RNA (mRNA) known as the cap – in a process known as 'cap snatching' – by binding to it and cleaving it off from the cell mRNA. The cap then becomes part of the virus' own mRNA, made by FluPol, so that the cell is 'fooled' into treating the viral mRNA as its own and thus 'translating' it into the viral proteins it needs to build new virus particles.
More information: Narin Hengrung et al. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus, Nature (2015).
Journal information: Nature
Provided by Diamond Light Source