Mathematical model gives insights into cotranscriptional folding of RNA
Researchers propose "Oritatami" as a mathematical model of the cotranscriptional folding of RNA in order to widen applications of RNA origami—one of the most significant experimental breakthroughs in molecular self-assembly.
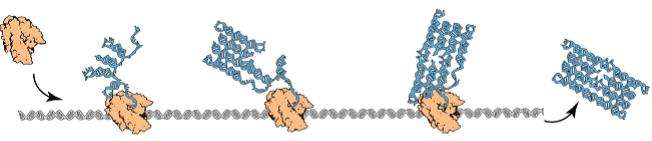
RNA origami by Geary, Rothemund, and Andersen is an experimental architecture of nanoscale rectangular tiles that self-assemble from an RNA sequence being folded cotranscriptionally, as shown in Figure 1. Theoretical models for programming this kind of molecular self-assembly are needed, as Winfree's abstract tile assembly model (aTAM) has yielded tremendous successes in experimental DNA tile self-assembly, which self-assemble a structure by letting unit DNA tiles attach to each other in some pre-programmed manner.
With Geary, now Shinnosuke Seki at the University of Electro-communications and his colleagues Pierre-Etienne Meunier and Nicolas Schabanel in Finland and France have proposed Oritatami to help understand the nature of cotranscriptional folding, the way that cells control RNA folding in vivo.
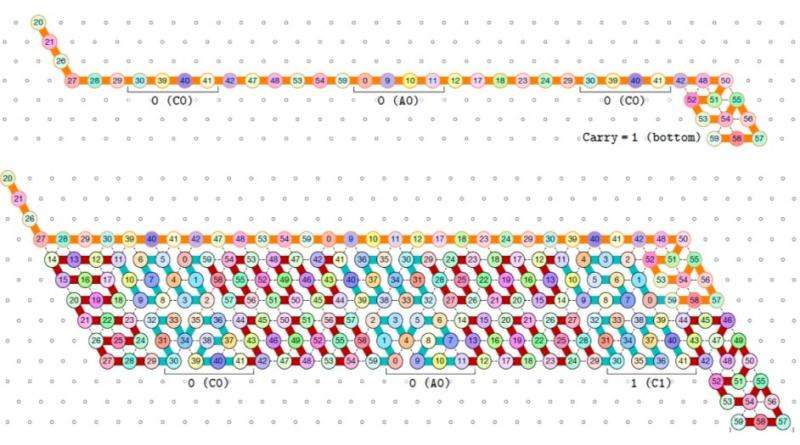
With this work, rather than simply predicting most likely conformations of RNA, they can implement computational devices out of RNA that take advantage of sequential folding to do something practical, such as count. The binary counter proof implements one of the most important types of device used in technology as a molecular self-assembly. The authors design an oritatami binary counter (see Figure 2), which suggests a way to use cotranscriptional folding for biomolecules to count in vivo. They also propose a fixed-parameter-tractable (FPT) algorithm to facilitate the design process of oritatami.
Transcription
Transcription is the first step of gene expression, in which a DNA template sequence, over A, C, G, T, is copied to an RNA transcript sequence, over A, C, G, U, called a messenger RNA (mRNA). An enzyme called RNA polymerase scans the DNA template and copies it letter by letter as A -> U, C -> G, G -> C, and T -> A.
Cotranscriptional folding
An RNA sequence tends to fold rapidly upon itself to take the most stable conformation, and hence, the RNA transcript already begins to fold while it is still being produced. What characterizes the folding of RNA transcripts is that transcripts fold in a continuous process while being transcribed. This means that the folding is controlled by the rate of strand production. This way of folding is hence called cotranscriptional or kinetic folding. In this way of folding, locally-stable structures of RNA will be preferred over some folds with better stability because they would require first unfolding parts of the strand in order to form.
Provided by University of Electro-communications