Co-evolution between a 'parasite gene' and its host
A Danish research team has delineated a complex symbiosis between a 'parasitic' noncoding RNA gene and its protein-coding 'host' gene in human cells. The study reveals how co-evolution of the host gene and parasite gene has shaped a feedback mechanism in which the parasite gene plays a completely new and surprising part as regulator of the host gene protein production. The breakthrough finding opens an entirely new avenue of research in gene expression.
There are many types of RNAs in human cells that do not act as a "recipe" for the production of proteins. These are called non-coding RNAs, and normally perform other jobs required for the functioning and general health of cells. The so-called small snoRNAs are located in the cell nucleus. These RNAs perform an important function as assistants in the production of other types of non-coding RNAs, more specifically the protein-producing factories known as ribosomes. The snoRNAs can guide the folding and maturation of the ribosomes by altering chemical groups in the ribosomal RNA. This job is carried out in collaboration with proteins that both help the snoRNA find the target RNA, and perform the chemical change at very specific positions.
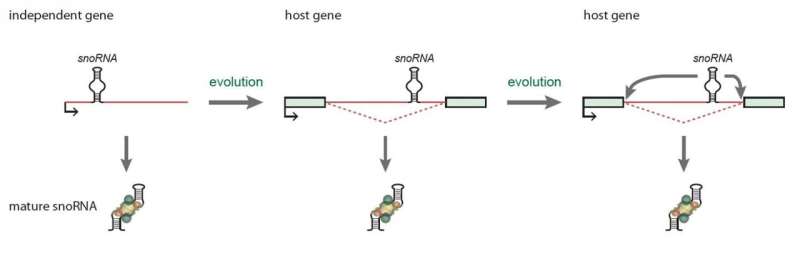
SnoRNA's function emerged early in evolution, and is found in all archaea and eukaryotic cells. Although their function and structure are preserved throughout evolution, the production of snoRNAs between cell types is very versatile. In humans, the majority of snoRNA genes are located within highly expressed protein-coding or non-coding host genes, and more precisely in parts of the host genes called introns; elements of the host gene that are excluded during the synthesis of the mature RNA during the process of splicing. This means that the snoRNA production is dependent on expression of the host gene.
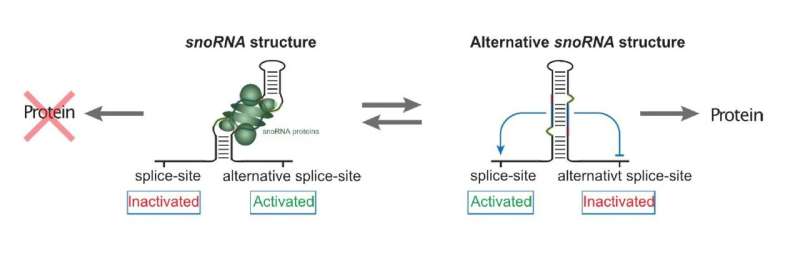
When the research team studied such snoRNA host genes, they identified a particular snoRNA that proved to have an alternative snoRNA-based task in the cell. They found that instead of assisting ribosomal production, this snoRNA acts as a sensor and master switch for the expression of the host gene, which encodes a snoRNA-binding protein necessary itself for the action of snoRNAs (Figure 1).
The results support a model in which, through structural changes, the snoRNA can regulate the splicing process of the host gene. At high snoRNA protein levels, the snoRNA structure will lead to an alternative splicing of the host gene's RNA, which will ultimately prevent further production of snoRNA protein. Conversely, lack of snoRNA proteins will lead to a different snoRNA structure during gene expression, resulting in increased snoRNA protein production.
Hence, the uncovered feedback mechanism ensures a precise coordination between snoRNA protein levels and global snoRNA levels, which ultimately ensures that other vital RNAs can be modified and produced properly. Misregulation of important non-coding RNAs is often associated with cancer and disease development, which underscores the need for a deeper understanding of the cell's strategies to maintain strict levels of functional snoRNA-protein complexes. In addition, these results strongly demonstrate that snoRNA parasitic genes located in host genes throughout evolution have enabled new and important roles, such as regulation of gene expression (Figure 2). This opens up a whole new research field where other snoRNA-regulated cellular mechanisms may be found.
More information: Søren Lykke-Andersen et al, Box C/D snoRNP Autoregulation by a cis-Acting snoRNA in the NOP56 Pre-mRNA, Molecular Cell (2018).
Journal information: Molecular Cell
Provided by Aarhus University














