RNA degradation contributes to gene silencing in higher eukaryotes
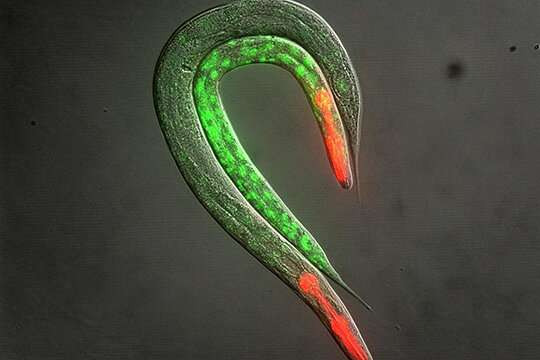
The Gasser group discovered that silencing of heterochromatic regions—and more specifically of hundreds of Polycomb-target genes enriched for H3K27me3—can occur through selective RNA degradation, and not only through transcriptional repression. The study links the epigenetics state of a gene with the fate of its RNA transcript. It is the first time that this is shown in higher eukaryotes.
Heterochromatin, the dark-staining transcriptionally silent compartment of the genome, correlates with repressed gene expression. In fission yeast and plants, RNA processing and degradation contribute to heterochromatin silencing, alongside conserved pathways of transcriptional repression. It has not been known whether similar pathways exist in higher eukaryotes where transcriptional repression is believed to be the main mechanism responsible for the silencing of genes in heterochromatin.
Ben Towbin, a former Gasser Ph.D. student, had already found some evidence that RNA degradation played a role in heterochromatin silencing in C. elegans: He identified three RNA binding proteins—all members of LSM complexes—that are involved in splicing and RNA decay, both in the cytoplasm and the cell nucleus. The C. elegans LSM proteins share 60-90% homology with the human LSMs, and assemble into two major complexes, LSM1-7 and LSM2-8.
Anna Mattout, a postdoc in the group of Susan Gasser, built on these findings in her own study in C. elegans, which has recently been published in Nature Cell Biology.
Having determined that it is the LSM2-8 complex that is involved in the repression of heterochromatin, Mattout looked for a link between this highly conserved RNA-binding complex and silencing mechanisms that work at the co-/post-transcriptional level. Combining microscopic, genetic and genomic approaches, she showed that LSM2-8 silences selectively heterochromatic reporters and hundreds of endogenous genes bearing almost exclusively the Polycomb mark H3K27me3. This acts by degrading selected transcripts through the XRN-2 exoribonuclease. Worms lacking the lsm-8 gene have developmental defects and die prematurely.
"The contribution of LSM8-mediated silencing of Polycomb-marked genes reveals a new mechanism and a new level of regulation that has not previously been demonstrated in higher eukaryotes," says Susan Gasser. "That mechanism has long been recognized to work at sites of constitutive heterochromatin bearing H3K9 methylation in fission yeast and plants. It appears that higher eukaryotes may have transferred the mechanism of post-transcriptional silencing to selectively degrade transcripts involved in development."
More information: Anna Mattout et al. LSM2-8 and XRN-2 contribute to the silencing of H3K27me3-marked genes through targeted RNA decay, Nature Cell Biology (2020).
Journal information: Nature Cell Biology