New AI-based versatile software for tracking many cells in 3D microscope videos

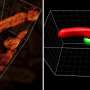
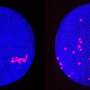
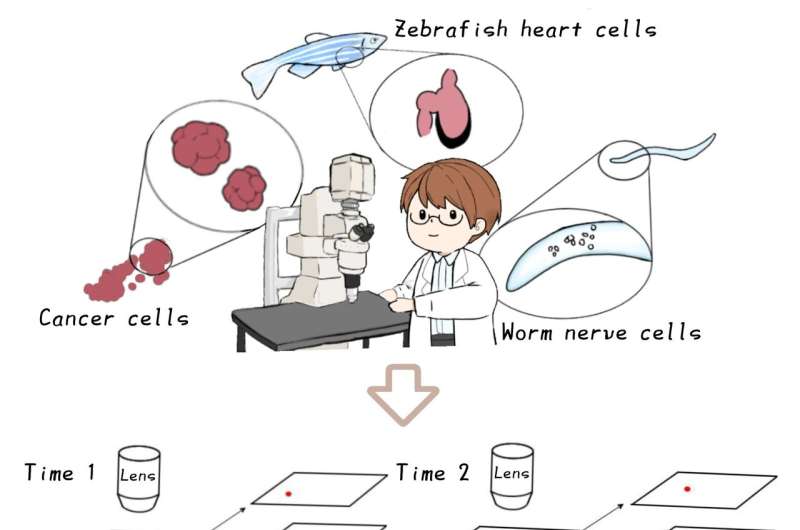
The first deep-learning software was developed as a versatile tool for tracking cells and extracting their signals from ~100 cells in a moving worm brain, in a zebrafish heart, and ~1,000 cultured cancer cells in 3D microscope videos. The method demonstrates significant improvements in tracking capabilities on various metrics including the possible number of tracked objects, robustness, and computing requirements.
In modern basic life science research as well as in drug discovery, recording and analyzing the images of cells over time using 3D microscopy has become extremely important. Once the images have been recorded, the same cell in different images at different time points has to be accurately identified (cell tracking) because the living cells captured in the images are in motion. However, tracking many cells automatically in 3D microscope videos has been considerably difficult.
In the Kimura laboratory at the Nagoya City University, Dr. Chentao Wen and colleagues developed the 1st AI-based software called 3DeeCellTracker that can run on a desktop PC and automatically track cells in 3D microscope videos. Using the software, they were able to measure and analyze the activities of ~100 cells in the brain of a moving microscopic worm, in a naturally beating heart of a young small fish, and ~1000 cancer cells cultured in 3D under laboratory conditions, which were recorded with different types of cutting-edge microscope systems.
This versatile software can now be used across biology, medical research, and drug development to help monitor cell activities.
More information: Chentao Wen et al, 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images, eLife (2021).
Journal information: eLife
Provided by Nagoya City University