Fluorescence lifetime imaging for studying DNA compaction and gene activities
Studies of the genomic DNA compaction in the cell nucleus and dynamic reorganization during physiologic processes or disease development in live cell environments are exceedingly challenging. This complexity stems from a high degree of compaction required to accommodate ~2 meters of genomic DNA within the cell nucleus, which typically is 5 to 10 micrometers in diameter. In addition, the chromatin compaction density is not static, but fluctuates over time accommodating for gene activities. Meanwhile, the 3D resolution of optical microscopy is not high enough even for sub-diffraction imaging modalities, thus limiting studies of the spatial geometry of complex genomic architecture and its dynamic transformations.
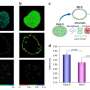
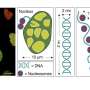
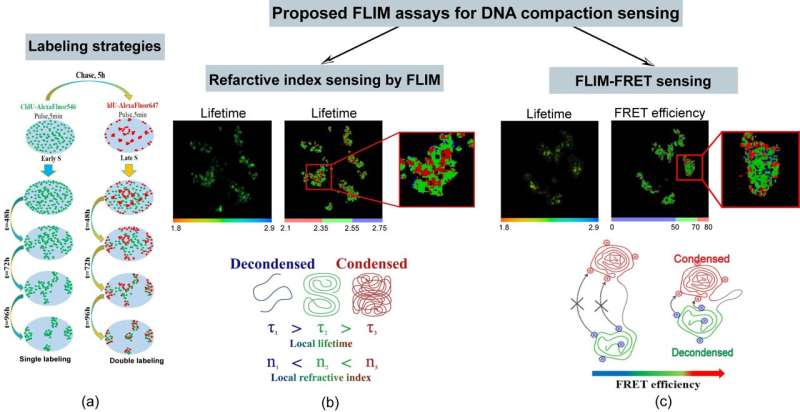
In a new paper published in Light: Science & Applications, a team of scientists, led by Professor Junle Qu from Center for Biomedical Optics and Photonics & College of Â鶹ÒùÔºics and Optoelectronic Engineering, Shenzhen University, China, and Prof. Paras N. Prasad from Institute of Lasers, Photonics and Biophotonics, State University of New York at Buffalo, U.S., has developed an alternative strategy that is based on fluorescence lifetime imaging (FLIM) to overcome existing limitations of conventional approaches. The authors propose two independent FLIM assays enabling fine measurements of DNA compaction. The first one relies on the inverse quadratic relation between the fluorescence lifetimes of fluorescent probes incorporated into DNA and their local refractive index, variable due to chromatin compaction density. Another FLIM approach employs Förster resonance energy transfer (FRET) between the fluorescence labeled nucleotides incorporated into the DNA strands.
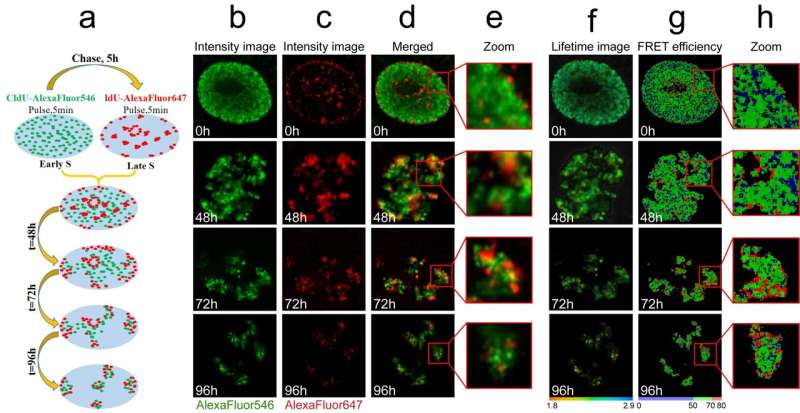
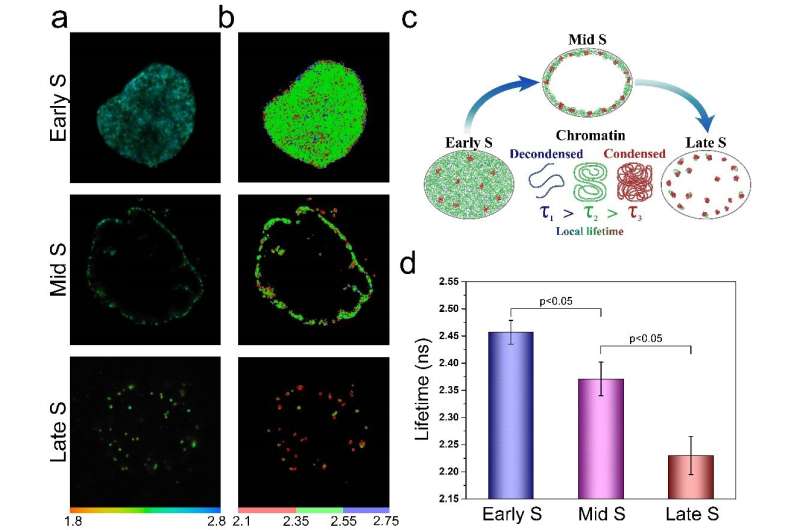
In this study, both FLIM assays were validated in cultured cells, where the researchers comparatively analyzed compaction of gene-rich chromatin domains that replicate in Early S-phase, and those that replicate in Mid- to Late S-phase and contain predominantly non-coding sequences. The obtained data demonstrate the sensitivity of both FLIM assays and unravel a significant difference in compaction of the gene-rich and gene-poor pools of genomic DNA. They show that the gene-rich DNA is loosely compacted compared to the dense DNA domains devoid of active genes.
More information: Svitlana M. Levchenko et al, Fluorescence lifetime imaging for studying DNA compaction and gene activities, Light: Science & Applications (2021).
Journal information: Light: Science & Applications
Provided by Chinese Academy of Sciences