April 14, 2023 feature
Ladder-shaped microfluidic systems for rapid antibiotic susceptibility testing

Thamarasee Jeewandara
contributing writer
The possibility of rapidly identifying antibiotic resistant bacteria can play a significant role in solving the global antibiotic crisis by facilitating the targeted and timely administration of pharmaceutical drugs. At present, the process of bacterial infection diagnostics take up to three days, which leads to less effective antibiotic treatment.
In a new report now published in Communications Engineering, Ann V. Nguyen and a research team at the Cornell University, New York, reported the development of a microfluidic system with a ladder-shaped design to generate a two-fold serial dilution of antibiotics, suited for national and international testing standards. The design and application process allowed them to scale-down the testing timeframe of antibiotic susceptibility to about 4–5 hours. The research outcomes were consistent with commercially available kits to provide an adaptable and efficient diagnostic tool for antibiotic susceptibility testing.
Antibiotic resistance and antibiotic susceptibility testing
The effects of antibiotic resistance are world-wide, and are credited to antibiotic overuse in human and veterinary medicine, and is associated with defects in . Researchers have noted the cause to be due to overprescribed antibiotics, and a lack of rapid laboratory tests and the circulation of prescription antibiotics that are only marginally effective. The capacity to develop and use rapid diagnostic tests can identify and characterize resistant bacteria to combat antibiotic resistance. The existing workflow of diagnosing antibacterial infection usually takes up to 2–3 days, during which a patient undergoes .
While the process begins with sample collection, biochemists can identify the infective agent depending on the characteristics of the pathogen, in a process that takes up to two days in a clinical microbiology lab. Researchers can isolate bacteria to conduct antibiotic susceptibility tests that take up another 16–20 hours resulting in a lack of timely acquisition of the information on .

Microfluidic platforms and the design of a microfluidic ladder-like instrument
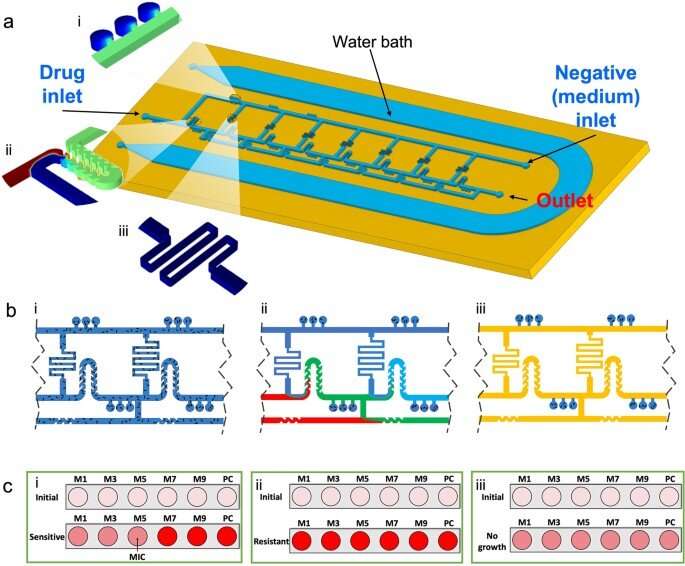
Bioengineers have therefore developed microfluidic platforms as a method to improve the sensitivity and speed of testing . In this work, Nguyen and the team designed and developed a microfluidic system with an optimized ladder-like network to combine and distribute culture media and antibiotics in a two-fold serial dilution. They studied the performance of the platform to conduct antibiotic testing, and developed an adaptable, diagnostic instrument. The setup included a microfluidic platform for antibiotic susceptibility testing, which combined nanoliter microchamber-based methods with a ladder-like concentration gradient generator.
The combination provided them with a standard and tunable antibiotic concentration profile to rapidly identify phenotypic antibacterial susceptibility. The researchers created the platform by incorporating a PDMS layer bonded to conventional glass slides, to allow them to test one antibiotic/bacterial combination per device. Each device served as bioreactors to incubate bacteria with antibiotics during the experiments.

The microfluidic instrument and its principle-of-action
The device contained three openings, including a drug inlet, a negative inlet, and an outlet to create a ladder-like structure. The team added the antibiotic and culture media to the device from opposing directions to dilute antibiotics as it moved across the system from the drug inlet, through a serpentine surface to dilute the solution and mix it with bacteria at specific flow rates. To identify the specific flow rate, the bioengineers calculated the required resistances and used computational fluid dynamics simulations to generate a greater range of dilutions. They then explored the principle-of-action of the microfluidic system with a .
The team used a cation-adjusted with PrestoBlue cell metabolism indicator in the setup. They then loaded the bacterial suspension into the device with a syringe followed by an antibiotic solution to generate a main channel network. They examined the performance of ladder microfluidic systems to determine the minimal inhibitory concentration of antimicrobial substances on the platform and examined bacterial isolates obtained from animal models. The instrument provided a framework to observe a variety of pathogens including Escherichia coli, and Staphylococcus pseudintermedius. In total, the bioengineers tested 206 bacterial samples on the microfluidic cell-sorting platform.

Outlook: Antibiotic susceptibility tests with spiked and clinical samples
Using the instruments, the team examined the possibility of bypassing the bacterial isolation step to conduct antibiotic susceptibility testing with urine samples. They accomplished this with urine samples cultured positive for the presence of a single unknown bacteria. The team determined the capacity for minimal inhibitory concentration from the accompanying clinical samples in 4–5 hours.
In this way, Ann V. Nguyen and colleagues designed and optimized a microfluidic system to perform phenotypic antibiotic susceptibility testing to isolate bacteria from culture plates or directly from urine samples. The instrument maintained a ladder-shaped architecture to generate a two-fold concentration gradient, which followed existing methods of standardization alongside clinically relevant antibiotics and drug combinations. The team conducted bacteria loading, antibiotic, and oil loading on the instrument and included a circuit logic for the first time in the study, to generate a microfluidics concentration gradient. The researchers plan to use the method to facilitate rapid antibiotic susceptibility testing to improve patient outcomes and streamline clinical laboratory workflow in a short timeframe.
The team also envisions integrating additional features to the ladder microfluidic instrument, including automatic loading capacity and sample handling within the ladder-chip for real-time image analysis with improved accuracy.
Written for you by our author —this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a (especially monthly). You'll get an ad-free account as a thank-you.
More information: Ann V. Nguyen et al, Ladder-shaped microfluidic system for rapid antibiotic susceptibility testing, Communications Engineering (2023).
Irith Wiegand et al, Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances, Nature Protocols (2008).
Journal information: Nature Protocols
© 2023 Science X Network



















