3D genome structure guides sperm development, shedding light on fertility and developmental disorders

Two new studies show how a seeming tangle of DNA is actually organized into a structure that coordinates thousands of genes to form a sperm cell. The work, published as two papers in Nature Structural & Molecular Biology, could improve treatment for fertility problems and developmental disorders.
"We are finding the 3D structure of the genome," said Satoshi Namekawa, professor of microbiology and molecular genetics at the University of California, Davis and senior author on one of the papers. "This is really showing us how the genomic architecture guides development."
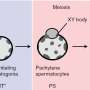
Although DNA is a long, stringy molecule, in living cells it is folded and looped like a ball of yarn. This means that genes can be physically close to the "enhancer" DNA switches that turn them on and off even if they are far apart in the DNA sequence.
To understand how genes are turned on and off to make different cell types, you have to figure out how the DNA is folded—and which genes and enhancers are paired together.
The memories of cells
In a mouse or human embryo, the cells that will one day produce sperm or eggs are already earmarked for that future purpose, Namekawa said.
These primordial germ cells are initially "bipotent"—able to become either sperm or eggs. But while the embryo is still snug in the womb, its bipotent cells commit to one path or another and once they cross that threshold, they cannot go back.

"Cells have a sort of memory," Namekawa said. "But we don't know how that memory works. We are trying to understand how this male fate is acquired."
Namekawa and his students used a technique called "Hi-C" to identify the proximity junctions where far-flung parts of the genome are paired up next to one another. Using a computer to analyze those paired locations, Namekawa was able to see how the entire string of DNA is looped and folded.
Bookmarking the genome
In the , UC Davis postdoctoral fellow Yuka Kitamura identified two proteins in germ cells that establish this cellular memory. The first, called SCML2, disconnects junctions and allows the DNA to unfold and loosen, preparing it to be reorganized in the next stage of sperm cell development.
Another protein, CTCF, attaches to locations of super-enhancer DNA and pairs them up with genes that they will later turn on during sperm development. This sets up a new structure that cements the cell's future fate as a sperm cell.

The shows that well before the germ cells enter meiosis, CTCF and other proteins bookmark thousands of locations in the genome. This paper was led by postdoctoral fellow Chongil Yi and Bradley Cairns, professor and chair of oncological sciences at the University of Utah School of Medicine and an investigator at the Howard Hughes Medical Institute. It was published in collaboration with Namekawa and Kitamura.
These discoveries could have important medical implications, including diagnostic tests for causes of infertility linked to genome folding.
The discoveries could also help scientists working on stem cell therapies, since coaxing a stem cell to become a neuron or heart cell requires shifting it from one genetic program to another—each defined by a particular 3D genome structure.
"We are uncovering the language of cell memory and cell fate," Namekawa said. "It's really exciting."
More information: Yuka Kitamura et al, CTCF-mediated 3D chromatin sets up the gene expression program in the male germline, Nature Structural & Molecular Biology (2025).
Chongil Yi et al, ZBTB16/PLZF regulates juvenile spermatogonial stem cell development through an extensive transcription factor poising network, Nature Structural & Molecular Biology (2025).
Journal information: Nature Structural & Molecular Biology
Provided by UC Davis