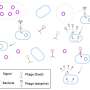
Fluorescence microscopy tracks phage attachment to bacteria in real time

Bacteriophages, or phages, viruses that selectively target and infect bacteria, have drawn growing attention for their potential use in a host of biotechnological processes to benefit humankind, from diagnosing contamination in consumer products to treating antibiotic-resistant infections.
But to achieve these advances, scientists must first know how phages specifically attach to bacterial cells early in infection—information that until now could be obtained only through a process that is labor-intensive and that yields limited insights.
In a recent study, however, Yale scientists describe a new method for quantifying the host-attachment dynamics of several phage species—including some that target key bacterial pathogens—offering a powerful tool for understanding these virus–cell interactions.
The new method, which utilizes fluorescence microscopy and particle tracking, in the Proceedings of the National Academy of Sciences.
"We decided to measure the attachment of individual viral particles to cells by directly visualizing them under the microscope," said Jyot Antani, an associate research scientist at Yale and lead author of the study. "Using automated particle tracking, we calculate the 'dwell time'—or the time that a phage spends interacting with bacterial cells—to measure phage attachment at single-virus resolution.
"Basically, we use a microscope to measure the 'stickiness' of phages to bacteria."
Antani is affiliated with the labs of Paul Turner, the Rachel Carson Professor of Ecology and Evolutionary Biology, and Thierry Emonet, the Lewis B. Cullman Professor of Molecular, Cellular and Developmental Biology, both in Yale's Faculty of Arts and Sciences as well as part of Yale's Quantitative Biology Institute. Turner and Emonet, along with Timothy Ward, a Yale undergraduate researcher, were also co-authors of the study.
Phages, which were first discovered more than a century ago, latch onto specific receptors on the surface of a bacterial cell and inject their genetic material into the cell to start the infection. Some phages, known as "lytic" phages, replicate themselves within host cells and destroy the cell by bursting it open, which helps control bacterial populations.
Phages are also ubiquitous: Scientists believe that phages, combined with the bacteria that they target, are the two most abundant biological groups on the planet, so their interactions have consequences that range from ecosystem function to community dynamics at the microbiome level.
Better understanding how they interact, the Yale researchers say, can help scientists predict the success of infection, improve the effectiveness of phage therapy, and guide the development of more efficient antibacterial strategies. It is important to understand factors at play early in the infection cycle, when the phage finds and binds to bacterial cell surface, Antani said. This key initial step is called "phage attachment."
When evaluating the relationship between phages and bacteria, scientists typically use what is known as the adsorption assay method, which calculates the rate of attachment over time. Specifically, the process involves mixing bacteria and phages together and then counting the number of "free" phages depleting over time to calculate an adsorption rate.
Unfortunately, Antani said, this method usually is only able to calculate attachment at population levels. It also requires large amounts of supplies and materials to grow the host bacteria, ample human labor, and a slow incubation process—and, even then, yields only a population-average readout.
In the new method, a fluorescent dye is used to label different viruses, and a camera records video of their interactions with bacteria immobilized on a glass coverslip surface. Using automated particle tracking, the researchers were able to obtain X–Y trajectories of individual phages.
"We noted significant variation in these trajectories' durations [dwell time], highlighting heterogeneity in viral particle attachment," Antani said. "By calculating the average dwell time and comparing it with traditional adsorption rate constants, we confirmed that our single-virus measurements closely match traditional bulk measurements, thus validating our new method."
Based on these findings, the new method offers promise for a wide range of uses, researchers say. For instance, they envision it being used to identify the best-binding phages for specific bacterial strains, including perhaps from patient samples.
The ability to achieve improved phage specificity could help identify bacterial species from samples in clinical and ecological environments, Antani said. "We aspire to develop a portable device for on-site diagnosis and are seeking industry partnerships to accelerate this process," he added.
"Our goal is to continue working with Yale Ventures to explore the possibility of licensing this technology, and we are excited to present on this single-particle attachment method at the upcoming Yale Innovation Summit," Turner explained. "We see the opportunity for developing other methods for measuring traits of individual virus particles—for example, when phages interact with bacterial host cells during stages of replication after the attachment process."
More information: Jyot D. Antani et al, Microscopic phage adsorption assay: High-throughput quantification of virus particle attachment to host bacterial cells, Proceedings of the National Academy of Sciences (2024).
Journal information: Proceedings of the National Academy of Sciences
Provided by Yale University