Interactive map of the human cell reveals previously unknown protein functions

Scientists have attempted to map the human cell since the first microscope was invented more than 400 years ago. But many components of the cell still remain uncharted.
" We know each of the proteins that exist in our cells, but how they fit together to then carry out the function of a cell still remains largely unknown across cell types," said Leah Schaffer, Ph.D., a postdoctoral research scholar at UC San Diego School of Medicine.
Now, Schaffer and her colleagues at UC San Diego—in collaboration with researchers at Stanford University, Harvard Medical School and the University of British Columbia—have created a comprehensive, interactive , which are associated with pediatric bone tumors. The study was in Nature.
They combined high-resolution microscope imaging and biophysical interactions of proteins to map the subcellular architecture and protein assemblies in the cell.
The map revealed previously unknown protein functions and will help the researchers understand how mutated proteins contribute to diseases such as childhood cancers. It will also serve as a reference for developing maps of other cell types.
"Based on cell biology 101 and textbook pictures of cells, you might think that we understand everything about a cell. But what's remarkable is that for no human cell type do we really have a proper parts catalog and assembly manual," said co-senior author Trey Ideker, Ph.D., a professor of medicine, adjunct professor in Jacobs School of Engineering and member of Moores Cancer Center at UC San Diego.
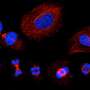
The researchers used a technique called affinity purification to isolate individual proteins and document their interactions with other proteins. In addition, they analyzed more than 20,000 images of the interior of cells marked with fluorescent dye to light up the location of proteins of interest from the .
Combining these data for more than 5,100 proteins revealed 275 distinct protein assemblies of different sizes within U2OS cells.
"Historically, scientists have been biased by the notion that one gene codes for one protein that has one function," said co-senior author Emma Lundberg, Ph.D., associate professor of bioengineering and of pathology at Stanford University.
"However, there is now an increasing number of known multifunctional proteins, and while we're probably still underestimating how many there are, this study demonstrates the importance of multimodal data integration to reveal these multifunctional properties."
The researchers discovered 975 previously unknown functions for proteins in the map. For example, C18orf21—a recently discovered protein whose function was previously unknown—appears to be involved with RNA processing, according to the study, and the DPP9 protein, known to cut proteins at specific regions, is implicated in interferon signaling, which is important for fighting infection.
The model drew upon a huge knowledge base it absorbed from the scientific literature on proteins, according to co-first author Clara Hu, a biomedical sciences doctoral candidate in Ideker's lab. The researchers asked GPT-4—a large language model artificial intelligence tool similar to ChatGPT—for the function of individual proteins and how they worked together in protein assemblies.
This took a fraction of the time it would take a human researcher, says Hu. This , published in Nature Methods, summarized the common theme of each protein assembly and proposed names for them, which were used in the cell map.

"We're able to, in an unbiased manner, really look at how these parts fit together and how to look at them in the context of disease," said Schaffer.
In fact, by locating mutated proteins on the cell map, the researchers were able to identify 21 assemblies frequently mutated in childhood cancer. Within these groups, 102 mutated proteins were found to be strongly linked to cancer development, thanks to the study. The findings have implications for how cancer research is conducted at the molecular and cellular level.
"We need to stop looking at the level of individual mutations, which are very rare, sporadic, and almost never recur in the same way twice, and start looking at the common machinery inside of cells that is disrupted or hijacked by these mutations," said Ideker.
Schaffer says browsing the U2OS cell map is similar to navigating an online geographical map.
"You're able to really explore, zoom in, and see what proteins are part of these different communities, and then see where those communities are located," she said.
"As you increase resolution, you can see even more detail-level information," said Hu. The team is currently working on resolving the map even further so that users can zoom in as much as they want at a high resolution.
The researchers think the U2OS cell atlas will not only facilitate a better understanding of childhood cancers, but will also provide a blueprint for scientists who want to map other cell types, use artificial intelligence tools to uncover the function of poorly understood proteins and protein complexes, and decipher the mechanisms behind a wide variety of disease processes.
More information: Trey Ideker, Multimodal cell maps as a foundation for structural and functional genomics, Nature (2025). .
Journal information: Nature Methods , Nature
Provided by University of California - San Diego