3D map of tongue's sweet sensor may lead to new ways to curb sugar cravings

Our attraction to sugar has grown to an unhealthy level. The average person in the United States now consumes , up from .
With published in Cell, Columbia University scientists have taken a major step toward dealing with this public health crisis. For the first time, they have mapped the 3D structure of the human sweet taste receptor, the molecular machine that allows us to taste sweet things. This could lead to the discovery of new regulators of the receptor that would significantly alter our attraction to and appetite for sugar.
"The leading role that sugar plays in obesity cannot be overlooked," said study co-first author Juen Zhang, Ph.D., a postdoctoral fellow in the lab of Charles Zuker, Ph.D., at Columbia's Zuckerman Institute and at the Howard Hughes Medical Institute. "The artificial sweeteners that we use today to replace sugar just don't meaningfully change our desire for sugar. Now that we know what the receptor looks like, we might be able to design something better."
The sweet receptors on our tongue can detect a large number of different chemicals that taste sweet, from common table sugar (also known as sucrose) to antimicrobial enzymes in chicken eggs. Unlike other receptors—for bitter, sour, or other tastes—our sweet sensors evolved to not be very sensitive. This helps us focus on sugar-rich foods for energy, and drives a need for a lot of sweets to satisfy our sweet tooth.
Determining the structure of the human sweet receptor is key to comprehending how it helps us detect sweet taste, fundamentally advancing our understanding of taste perception. More than 20 years ago, Dr. Zuker and his colleagues uncovered the genes behind .
This landmark work revealed its chemical formula, but until now no one knew its precise shape, much like how knowing a cake's recipe will not tell you what the pastry will look like when finished.
Without this knowledge, understanding the molecular basis of sweet detection to rationally design ways to regulate the function of this essential receptor has been a challenge, said Dr. Zuker, in whose laboratory this new work was also carried out.
"All the artificial sweeteners that we use today were either discovered by accident or based on known sweet-tasting molecules," said study co-author Brian Wang, a research assistant in the Zuker lab. "As a result, most artificial sweeteners have drawbacks."
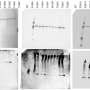
The new work maps the structure of the human sweet taste receptor in unprecedented detail, to a resolution as good as 2.8 angstroms. In comparison, the smallest atom, hydrogen, is slightly more than 1 angstrom wide.
It took the researchers innovative approaches and about three years to map the human sweet taste receptor's structure, in large part because it proved difficult to grow this protein on cells in lab dishes.
"Just getting the purified protein we needed to map the structure took more than 150 different preparations over the course of three years," said study co-first author Zhengyuan Lu, a doctoral student at the Zuker lab.
The scientists then used cryo-electron microscopy (cryo-EM) to analyze the human sweet taste receptor. This technique fires beams of electrons at molecules that have been frozen in solution, helping researchers capture snapshots of those molecules from different perspectives, from which they can reconstruct their three-dimensional structures at the atomic level.
Of particular importance, cryo-EM revealed the receptor's binding pocket: the cavity where sweet things stick and trigger the set of reactions that drive our strong appetite for sweets.
"Defining the binding pocket of this receptor very accurately is absolutely vital to understanding its function," said study co-author Anthony Fitzpatrick, Ph.D., a principal investigator at Columbia's Zuckerman Institute.
"By knowing its exact shape, we can see why sweeteners attach to it, and how to make or find better molecules that activate the receptor or regulate its function," added Dr. Fitzpatrick, who is also an assistant professor of biochemistry and molecular biophysics at Columbia's Vagelos College of Â鶹ÒùÔºicians and Surgeons.
The human sweet taste receptor consists of two main halves. One of these, named TAS1R2, possesses the binding pocket, a component resembling a Venus flytrap. Knowing the structure of this part may also help us understand why people differ in how sensitive they are to sweets.
The scientists mapped the receptor's structure as it bound to two of the most commonly used artificial sweeteners, aspartame and sucralose. These are, respectively, 200 and 600 times sweeter than sucrose.
The researchers then systematically altered tiny parts of the receptor. This helped shed light on the role each of these parts play in binding onto the sweeteners, said study co-author Ruihuan Yu, a doctoral student at the Zuker lab.
"We're trying to move our understanding of science forward to be able to help people," said study co-author Andrew Chang, a research technician at the Fitzpatrick lab.
Although the human sweet taste receptor is found mostly on taste buds in the mouth, Dr. Zhang noted it is also scattered throughout the body, where it may play a role in the function of organs such as the pancreas.
As such, the new map of this receptor's structure might support research investigating our metabolism, as well as in disorders such as diabetes.
More information: The Structure of Human Sweetness, Cell (2025). .
Journal information: Cell
Provided by Columbia University