Mass spectrometry method identifies pathogens within minutes instead of days

Traditionally, bacterial diseases are diagnosed by the tedious isolation of pathogens and the creation of bacterial cultures. Waiting times of several days are the rule here. Only then can targeted treatment of the disease begin.
Researchers at the Technical University of Munich (TUM) and Imperial College London have developed a new method to identify bacteria with unprecedented speed. This means that the waiting time can be reduced from several days to just a few minutes.
The work is in the journal Nature Communications.
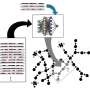
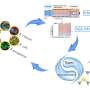
The team, led by Nicole Strittmatter, professor of analytical chemistry at TUM, and Dr. James S. McKenzie (Imperial), uses mass spectrometry for its innovative approach. This enabled the researchers to identify specific metabolic products of bacteria directly in tissue and stool samples.
At the heart of the process is a database in which 232 medically important bacterial species and their metabolic products have been recorded to date. Biomarkers are derived from this database, which can then be used to directly detect specific bacteria.
Among the bacteria that can be identified using the new method are clinically extremely important pathogens that can, for example, trigger stomach cancer, are responsible for certain pneumonias and meningitis, are associated with premature births, and can cause gonorrhea or blood poisoning.

Further expanding the bacterial database
First author Wei Chen, Ph.D. student at the Department of Bioscience at the TUM School of Natural Sciences in Garching, emphasizes, "Our innovative approach is not to look directly for the pathogenic bacteria, but only for their metabolic products. This allows us to detect them indirectly, but much more quickly."
Prof. Nicole Strittmatter also sees great opportunities for use in personalized medicine, in which the therapy is precisely tailored to the respective patient. "This is one of the most important future topics in biotechnology and medicine. Targeted interventions can dramatically improve the chances of successful treatment. As analysts, we develop modern tools and methods for doctors to do this."
The biomarker database now needs to be further expanded to enable the regular use of the new method in clinical practice. According to the researchers, a total of over 1,400 bacterial pathogens are known and described. Their specific metabolic products should now be identified and included.
More information: Wei Chen et al, Universal, untargeted detection of bacteria in tissues using metabolomics workflows, Nature Communications (2025).
Journal information: Nature Communications
Provided by Technical University Munich