New method allows live-cell protein tagging without interfering with function

University of Pennsylvania researchers have developed a powerful new investigative tool to reveal how proteins work inside living cells. A technique that enables quick, precise and minimally disruptive incorporation of amino acids and chemical labels into proteins within live mammalian cells.
Studying proteins in their live cellular context is essential for understanding biological function. Genetically encoded tags, including fluorescent proteins and epitope sequences, are widely used to monitor protein behavior.
Large tags can alter protein localization or disrupt native interactions. Small-molecule labels offer less structural interference yet remain difficult to apply within living cells. Antibodies serve as another approach yet are often incompatible with live-cell experiments due to their size and specificity.
Most existing techniques lack temporal precision and fail to capture protein activity on timescales relevant to cellular signaling and adaptation. A technology that enables precise, minimally invasive, and rapid labeling of proteins under native expression conditions remains unmet.
In the study, "Intracellular protein editing enables incorporation of noncanonical residues in endogenous proteins," in Science, researchers introduce a method for editing protein sequences inside living mammalian cells to site-specifically incorporate chemically modified amino acids, epitope tags, or other peptide segments into endogenous or exogenously expressed proteins with temporal control.
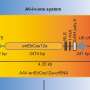
Researchers developed a protein editing mechanism using two engineered molecular tools that act like programmable splicing switches. When a donor protein enters the cell, these tools activate and precisely insert a chosen protein segment into a specific position within the target.
Inserted segments are joined seamlessly while the molecular machinery that enabled the insertion is removed. Editing can either insert new features or restore original ones, depending on how the system is configured.
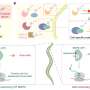
Protein editing was first demonstrated using model proteins including calnexin and β-actin. Successful insertion of new amino acid sequences into these proteins was confirmed using immunoblotting, microscopy, and mass spectrometry. Editing took place within 10 minutes of delivering the donor protein into the cell, offering a level of speed not typically achievable with other tagging methods.
In further tests, the method was applied to proteins with essential cellular functions, including the transcription factor c-Myc and the kinase Chk1. Specific protein segments were temporarily replaced with identifiable tags and then restored to their original sequences. After editing, both proteins retained normal activity, as shown by phosphorylation events and preserved DNA-binding interactions.
Researchers also demonstrated that edited proteins could carry small molecules such as fluorophores or biotin by modifying the donor protein to contain a synthetic amino acid with a chemical handle.
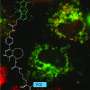
After labeling these in vitro, the modified proteins were delivered into mammalian cells and spliced into targets such as β-actin or endogenous calnexin. Labeled proteins retained correct cellular localization and were trackable using confocal microscopy or isolated using affinity purification.
Lipid nanoparticles were tested as an alternative to electroporation for protein delivery. Editing was successful using both methods, although lipid nanoparticles were especially effective when donor proteins were engineered with a negatively charged tail to aid delivery.
Fluorescent signals from edited proteins persisted for hours and matched the expected location of the target protein inside the cell. Calnexin, for instance, is localized to the endoplasmic reticulum, while β-actin is assembled in the cytoskeleton. Signals from unedited or improperly delivered proteins faded quickly, suggesting minimal background interference. Splicing occurred within minutes, enabling access to protein populations on biologically relevant timescales.
Potential applications of the technique span molecular cell biology with implications for disease research, drug development, and biotechnology.
More information: Jenna N. Beyer et al, Intracellular protein editing enables incorporation of noncanonical residues in endogenous proteins, Science (2025). .
Journal information: Science
© 2025 Science X Network