Study reveals intricate molecular detail of human DNA repair process

Lisa Lock
scientific editor

Robert Egan
associate editor

Researchers have revealed the structural mechanisms of a major DNA repair pathway in human cells.
The research, today as a Reviewed Preprint in eLife, is described by the editors as a landmark study with compelling evidence on how an important player in DNA repair—the RAD51 filament—promotes the exchange of strands between DNA molecules that contain homologous (identical or similar) sequences. They added that the findings will be very valuable for research communities studying DNA repair and genome stability.
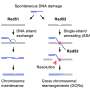
Homologous recombination (HR) is one of the key DNA repair pathways in cells. It is essential for repairing double-stranded breaks in DNA and for DNA crossover events during sexual reproduction. Moreover, cells deficient in HR are more prone to cancer, and targeting the cells' HR machinery—together with other DNA repair pathways—can be used to kill cancer cells (an approach called synthetic lethality).
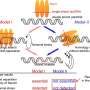
A unique feature of HR is that it repairs the damage to the DNA in a highly accurate way. In eukaryotic cells (those with a nucleus), HR is performed by an enzyme called RAD51, which binds to single-stranded DNA at the site of damage, forming a nucleoprotein filament.
This filament performs a physical search for a complementary sequence by "invading" a sister chromatid or homologous chromosome. Once the correct sequence is found, the filament creates a stable three-stranded structure called a displacement loop (or D-loop) and the process of 'strand exchange' occurs, but the mechanistic understanding of this last step remains limited.
"We have obtained a cryo-electron microscopy structure that captures the state of human Rad51 tightly bound to broken DNA," says co-lead author Luay Joudeh, Microscopy Manager at the Departments of Pathology & Biochemistry, University of Cambridge, U.K. "The structure reveals for the first time the mechanism underlying the strand exchange process during DNA repair via homologous recombination."
Joudeh served as co-lead author of the study alongside Robert Appleby, who was a Ph.D. student in the Department of Biochemistry, University of Cambridge, at the time of the study, and is now a Postdoctoral Fellow at the Francis Crick Institute, London, U.K.
The team began by preparing a D-loop sample of human RAD51 in the laboratory that could be visualized using a powerful technique called cryo-electron microscopy (cryoEM). They collected thousands of movies of the D-loop sample, which allowed them to construct a final 3D structure from the individual images of more than 100,000 physical D-loop particles. This structure revealed the position of each HR component during the strand exchange process.
"We found that, to search for the complementary sequence to use in the repair, RAD51 first unwinds the double helix of the donor DNA by inserting physical spacers to separate the unwoven strands," explains senior author Luca Pellegrini, Professor of Structural Biology in the Department of Biochemistry, University of Cambridge.
These physical spacers are known as L2 loops, and each insertion exposes the bases of three DNA nucleotides that are then available for the invading strand to pair to, probing for homology.
Pellegrini continues, "One interesting observation we made is that at one end of the D-loop structure, the L2 loops of RAD51 cause a large alteration to open up the donor DNA, but at the other end of the D-loop they play a much more limited role. This indicates a preferred orientation in the process of D-loop formation and shows that the L2 loops have an inherent flexibility that enables them to behave differently along their interface with the target DNA."
Other surprising discoveries for the team included the finding that the invading strand binds to the entire complementary sequence of the donor DNA except for one available base pair that remains unfulfilled. The authors speculate that unwinding of the DNA during HR increases the distance between the two nucleotides, preventing them from being able to pair.
At the other end of the complementary sequence, one nucleotide mismatch (pairing that doesn't follow the Watson-Crick base pairing scheme) between invading and donor strands shows that D-loop formation can occasionally tolerate differences in sequence.
Finally, the cryoEM structure revealed that the exchanged strand is captured in a channel within the RAD51 complex that facilitates its removal. They were also able to identify the exact components of RAD51 that create this channel and how they are positioned along the DNA strands.
"Together, these findings form the basis of a model of HR in which RAD51 binding directs the broken DNA into a position where the physical spacers (the L2 loops) can separate the strands, capture the exchanged strand and facilitate strand exchange," explains Appleby.
"Our RAD51 D-loop structure provides fundamental insights into the biochemical reaction of eukaryotic homologous recombination," says Pellegrini. "We believe these insights will be useful for those studying DNA repair processes in health and disease, and in designing therapies that can either repair or block these mechanisms in diseases such as cancer."
More information: Luay Joudeh et al, Structural mechanism of strand exchange by the RAD51 filament, eLife (2025).
Journal information: eLife
Provided by eLife