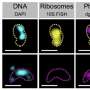
Strategy to delimit flexible regions within the SAR11-gMED-CCG. Credit: Microbiome (2025). DOI: 10.1186/s40168-025-02037-6
Using third-generation metagenomics, researchers from the Department of Plant Production and Microbiology at the Miguel Hernández University of Elche (UMH) have uncovered the genomic basis behind the evolutionary success of the SAR11 marine bacteria, the most abundant bacteria in the ocean.
Published in the journal Microbiome, reveals that these bacteria combine a shared "genetic core" with small genomic islands, each typically harboring a single "flexible gene" that allows the population to respond to environmental changes dynamically.
This discovery sheds light on how these microbial populations are essential to maintaining global ecological balance, fostering biodiversity, and adapting to the impacts of climate change.
The marine microbiome plays a crucial role in sustaining ecosystems, driving global biogeochemical cycles, and accounting for up to 98% of marine primary productivity. SAR11 is a clade of free-living bacteria that numerically dominates ocean surface waters, making up 20–40% of all prokaryotic cells.
"Despite the abundance and cosmopolitan distribution of these microbes, limitations in recovering the full genetic richness of their natural populations have hindered our understanding of the link between microbial evolution and ecology from a genomic standpoint," explains Mario López Pérez, UMH researcher and senior corresponding author of the study.
For the first time, the UMH Microbial Genomics and Evolution Group has combined single-cell genomics and long-read metagenomics to accurately reconstruct the genetic diversity of SAR11 in environmental samples from the Mediterranean Sea. This innovative approach made it possible to unravel how the SAR11 genome is structured and how the strains coexisting in the same population diversify.
The study shows that these bacteria share an almost identical genetic core, representing 81% of their genome. The remaining portion, known as the flexible genome, is concentrated in small regions, most containing a single gene, and appears in equivalent positions across all strains.
The image shows the 'phylogenetic tree' of a group of genes called Amt, found in SAR11 bacteria. Different versions of these genes, which may influence how the bacteria take up and process nitrogen, are shown in different colors. Brown branches indicate core versions to the entire population, while green dots mark variants found in Mediterranean strains. The protein structures are also overlapped, highlighting that they all retain similar functions despite their differences. This genetic variation enables these bacteria to adapt to different marine environments. Credit: Molina-Pardines et al., 2025, Microbiome. DOI: 10.1186/s40168-025-02037-6
"These small variations are always in the same location within the genome and contain genes with similar functions, although in different versions," explains Carmen Molina Pardines, UMH Ph.D. student and first author of the study.
This genomic pattern favors the coexistence of multiple strains and minimizes direct competition among them.
This organizational structure gives rise to polyclonal populations—groups of multiple genetic variants coexisting in the same environment.
Such a structure preserves essential genes during selective sweeps and maintains functional redundancy, safeguarding a broad genetic reservoir. This genetic reservoir allows the population to adapt to environmental pressures rapidly.
"These results offer insight into the strategies that explain the ecological success of SAR11 in nutrient-poor marine environments like the Mediterranean," adds José M. Haro Moreno, UMH researcher and co–primary author of the study.
Beyond its evolutionary insights, the study also demonstrates that third-generation metagenomics overcomes technical limitations that previously made it difficult to study these microorganisms, which are extremely challenging to culture in isolation.
The study positions UMH as an international leader in marine microbiome evolution research. Its integrative approach contributes to a deeper understanding of the mechanisms that support life in the oceans—and, by extension, on Earth.
More information: Carmen Molina-Pardines et al, Extensive paralogism in the environmental pangenome: a key factor in the ecological success of natural SAR11 populations, Microbiome (2025).
Journal information: Microbiome
Provided by Miguel Hernandez University of Elche