Engineered enzyme enables detailed mapping of tRNA changes in tumor cells

Stephanie Baum
scientific editor

Robert Egan
associate editor

An engineered enzyme is at the center of a new method to visualize molecular details in human cells, and how these molecules change in cancerous versus benign cells, Boston College researchers in the journal Cell Chemical Biology.
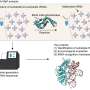
The method—dubbed "MapID-tRNA-seq"—allowed researchers to examine all tRNAs and their changes within human cells at once, according to the report. The study focused on transfer RNAs, or tRNAs, which ferry protein building blocks inside of cells, making them biologically essential. Dysregulated tRNAs are frequently involved in human diseases.
"Our new method helped reveal critical changes in tRNAs that occur during diseases, specifically, cancerous versus benign breast tumor cells, according to Boston College Assistant Professor of Chemistry Huiqing (Jane) Zhou, who led the study, titled "MapID-based Quantitative Mapping of Chemical Modifications and Expression of Human Transfer RNA."
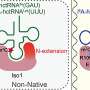
Zhou described tRNAs as very "bumpy" molecules for a cell to process. Most RNAs are composed of four nucleotides. However, tRNAs carry abundant chemical decorations on the nucleotides, which effectively block natural processing enzymes on tRNAs during these biological transactions.
"These events have largely impeded existing techniques to detect tRNAs," said Zhou. "We developed a method to effectively process human tRNAs by applying an engineered enzyme, which is a reverse transcriptase from our earlier work. This enzyme is critical in order to enable high-throughput and accurate detection of cellular tRNA molecules."
Zhou, joined in the project by graduate students Mitchel L. Tepe and Allison Carso, and undergraduate researcher Yitan Chen, studies human cells. Zhou's lab extracts and purifies molecules of interest, such as tRNAs, from cultured human cells and uses a next-generation-sequencing (NGS)-based approach to examine the molecular features of tRNAs.
Zhou and her team applied their method to four cell lines, including three mammary cell lines. Using MapID-tRNA-seq, the researchers were able to map both tRNA modifications and expressions, discovering that some modification profiles from cell to cell largely remain unchanged.
However, a few modification sites showed cell type specifics.
Zhou found that tRNA expressions are largely dysregulated, or altered, in cancerous mammary cell lines compared to the benign cells. The researchers compared one benign cell line to two cancerous tumor cell lines. The cancerous tumor cells showed significantly fewer mitochondrial-derived tRNAs compared to the benign tumor.
The team further explored mitochondrial expressions in these mammary cell lines and found that the ribosomal RNAs appeared to be less in cancerous tumor cells compared to the benign cell line, while messenger RNAs (or mRNAs) remain mostly unchanged. Other studies have shown that mitochondrial proteins are often downregulated.
"The role of mitochondria in cancer has been a focus of researchers. This leads us to believe that the gene expressions in mitochondria in breast cancer cells are in a defective state," said Zhou.
"Our data suggests that this protein dysregulation stems from the lack of tRNA and rRNA, which are crucial for protein production," said Zhou. "It is not from a lack of mRNA, or the blueprint of the proteins in question."
While mapping modifications in the cell lines, the team found new modification sites on tRNA that merit further study. One modification could directly impact protein translation during protein production, said Zhou. The team is at work trying to understand the function of the puzzling modification.
More information: Mitchel L. Tepe et al, MapID-based quantitative mapping of chemical modifications and expression of human transfer RNA, Cell Chemical Biology (2025).
Journal information: Cell Chemical Biology
Provided by Boston College