Whole plants of the wild rice allopolyploid Oryza minuta and its two diploid progenitors. Credit: Wang Qian
Polyploidy, or whole-genome duplication (WGD), is a major mechanism of genome evolution across the tree of life and is particularly prevalent in plants, where it facilitates the evolution of new traits.
Despite certain advantages, nascent polyploid plants face challenges at multiple levels—from molecular and cellular to ecological—due to the presence of an extra set of chromosomes. As a result, many polyploid plants undergo diploidization, a process that helps overcome these challenges and drives innovation in transcriptional, physiological and morphological traits.
Many so-called "diploid" plants are actually "paleopolyploids"—they underwent multiple rounds of ancient WGDs followed by subsequent diploidization process. This evolutionary cycle, which is widespread in plants, is known as the polyploidy–diploidization cycle.
Although this cycle is common, the tempo and mechanism of diploidization have not been well understood, primarily because it is difficult to distinguish between de novo evolution (new changes following polyploidy) and genetic variation inherited from parental species.
Now, however, a research team led by Prof. Ge Song from the Institute of Botany of the Chinese Academy of Sciences, in collaboration with partners, has investigated the diploidization process in the wild rice allotetraploid, Oryza minuta, using a population genomic approach. Their findings reveal that the tempo of diploidization within a single polyploid species can be both episodic and gradual. The results are published in .
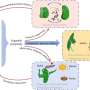
Exploration of impact of DNA methylation, TE distribution, and parental legacy on the expression patterns of homoeologous genes in Oryza minuta. Credit: Wang Xin & Zou Xinhui
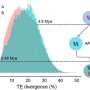
The researchers generated chromosome-scale genome assemblies for three wild rice species. Through extensive population sampling, they successfully separated inherited variation from post-polyploidy evolution, uncovering a nuanced picture of diploidization: whereas gene fractionation (the loss of duplicated genes) occurred in an early burst, the accumulation of transposable elements (TEs) and homoeologous exchanges proceeded gradually.
The patterns of homoeolog expression bias—the preferential expression of one copy of the duplicated genes over the other—were highly variable across different tissues, with no consistent subgenome bias. The researchers further evaluated the roles of DNA methylation, TE distribution, and parental legacy in shaping expression patterns, highlighting the complexity of transcriptional regulation.
By showing that diploidization in a single polyploid species can be both episodic and gradual, depending on the type of mutation, the study underscores that "diploidization" is a heterogeneous genomic process.
The findings not only deepen our understanding of plant genome evolution but also demonstrate the power of population genomics to reveal the dynamics and mechanisms driving polyploid evolution.
More information: Xin Wang et al, Diploidization in a wild rice allopolyploid is both episodic and gradual, Proceedings of the National Academy of Sciences (2025).
Journal information: Proceedings of the National Academy of Sciences
Provided by Chinese Academy of Sciences