High-speed AFM and 3D modeling reveal the dynamics of a protein implicated in several cancers

Lisa Lock
scientific editor

Robert Egan
associate editor

An enzyme type noted in several cancers is the family of adenosine deaminases acting on RNA (ADARs). These enzymes convert adenosines in double-stranded RNA (dsRNA) into inosines, which cells read as guanosines. As such, ADARs can contribute to changes in protein-coding sequences and diminish the robustness of various RNA processes.
Studies have shown that silencing one type of ADAR—ADAR1—can prevent cancer proliferation and sensitize cancers to immunotherapy, suggesting that they could be a promising target for cancer treatments.
However, it has so far been difficult to pin down information on the structural dynamics of ADAR1 due to its size and complexity. Now, researchers have combined high-speed atomic force microscopy (HS-AFM) and 3D modeling to shed light on the enzyme's conformations and interactions with dsRNA.
The team was led by Madhu Biyani at Kanazawa University, WPI-NanoLSI, Yasuhiro Isogai at Toyama Prefectural University, and Manish Biyani at Ishikawa Create Labo and Kwansei Gakuin University. Their findings are in Nature Communications.
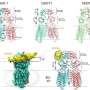
Like many proteins, ADAR1 functions through changes in its conformation. However, most experimental techniques for determining protein structure, as well as 3D modeling algorithms, give static or average conformations that obscure the structural dynamics so important to the protein's function. Combining 3D modeling with HS-AFM proved helpful in shedding light on these dynamic aspects of ADAR1.
The researchers first used 3D modeling based on the machine learning algorithm AlphaFold2 to predict the conformations of the enzyme and noted that it could take the form of monomers, dimers, trimers, and tetramers. HS AFM observations, as well as theoretically simulated HS AFM, supported these initial conclusions regarding the possible oligomer formations.
The researchers then looked at the conformations the enzyme formed in the presence of double-stranded RNA (dsRNA). In particular, the researchers focused on a certain aryl hydrocarbon receptor 3'UTR mRNA as the target for ADAR1, since this receptor is known to be involved in the metabolism of substances alien to the body at those points. Observations of the dsRNA with HS-AFM not only agreed with previous structural studies but were able to provide insights into the structure of the target region in particular.
Thanks to the speed and resolution of the HS-AFM image capture, the researchers were able to identify different conformations in the proteins that seemed to relate to distinct phases of the deaminizing process.
In the study, the researchers explain how ADAR1 first searches for the dsRNA and on recognizing it, adopts a flexible conformation as it approaches. The enzyme then engages in what the researchers describe as "capture" of the dsRNA backbone, for which the conformation transitions to something more stable and anchor-like.
The researchers highlight the role of dsRNA binding domains (dsRBDs) to stabilize the interaction with the dsRNA at this stage. They also note "a visibly large interfacial interaction between the deaminase domains, forming a dimer" as the enzyme dimer loops out on the dsRNA. The enzyme subsequently scans the RNA and dissociates to search for adenosine sites to convert.
"These observations suggest that the dsRBDs are critical for initiating interactions between the deaminase domains, thereby promoting the formation of a stable, functional dimeric complex capable of efficiently binding and catalyzing the editing of dsRNA substrates," the researchers conclude in their report, thereby flagging the insights this study offers for further work towards possible cancer therapeutics.
The researchers further propose future studies to compare ADAR1 and ADAR2, and to perform mutation analyses to clarify how ADAR1 dimerization influences A-to-I RNA editing, ultimately aiming to develop effective ADAR1 inhibitors.
More information: Madhu Biyani et al, High-speed atomic force microscopy and 3D modeling reveal the structural dynamics of ADAR1 complexes, Nature Communications (2025).
Journal information: Nature Communications
Provided by Kanazawa University