Intestinal bacterium allows microbiome-mediated protection against pathogens

Gaby Clark
scientific editor

Robert Egan
associate editor

The totality of bacteria, viruses and fungi that exist in and on a multicellular organism forms its natural microbiome. The interactions between the body and these microorganisms significantly influence both, the functions and health of the host organism. Researchers assume that the microbiome plays an important role in the defense against pathogens, among other things.
The Collaborative Research Center (CRC) 1182 "Origin and Function of Metaorganisms" at Kiel University has been investigating the highly complex interplay between host organisms and microorganisms for several years using various model organisms, including the nematode Caenorhabditis elegans.
In a recent study, researchers from the CRC 1182 have gained new insights into the molecular mechanisms within the microbiome which contribute to the defense against pathogens. In collaboration with scientists from the Max Planck Institute for Terrestrial Microbiology and the University of Edinburgh, they discovered that a protective bacterium of the genus Pseudomonas, which is found in the intestinal microbiome of C. elegans, produces sphingolipids.
This result was surprising, as it was previously assumed that the production of sphingolipids was restricted to only a few bacterial phyla and the bacterial genus Pseudomonas was not known to be able to produce these specific molecules.
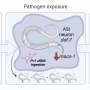
The researchers discovered that Pseudomonas utilizes an alternative metabolic pathway for sphingolipid production, which differs significantly from the known sphingolipid synthesis pathways in other bacteria. They were also able to show that the sphingolipids produced by Pseudomonas bacteria play an essential role in protecting the intestinal epithelium of the host from damage by the pathogen.
Responsible for sphingolipid production in Pseudomonas bacteria is a specific biosynthetic gene cluster that forms the enzymes for this novel metabolic pathway. Interestingly, similar gene clusters were also found in other host-associated gut bacteria, suggesting that the ability to produce sphingolipids may be more widespread than previously thought. This suggests that bacterial sphingolipids may play a central role in microbiome-mediated protection against infection—not only in C. elegans, but potentially also in other host organisms.
The results of the interdisciplinary study, conducted under the leadership of PD Dr. Katja Dierking (Evolutionary Ecology and Genetics research group at Kiel University), in collaboration with other research groups from Kiel and national and international cooperation partners, were recently in the journal Nature Communications.
In 2019, the Kiel research group published a in Current Biology that showed that certain members of the C. elegans microbiota protect against pathogen infection. "For one Pseudomonas species, we knew that it could protect the worm from infections. However, we had not yet been able to identify the substances and mechanisms involved," emphasizes Dr. Lena Peters, a scientist in the Evolutionary Ecology and Genetics research group.
In a broad-based collaboration of scientists both within the CRC 1182—including Kiel professors Christoph Kaleta and Manuel Liebeke—and with external scientists, including Professor Helge Bode from the MPI for Terrestrial Microbiology in Marburg and Professor Dominic Campopiano from the University of Edinburgh in Scotland, the genetic and metabolic basis of the protection against infection mediated by the microbiome was analyzed.
Using metabolic and transcriptional studies, single-molecule analyses and mass spectrometry approaches, the researchers made a surprising discovery: They were able to prove that the protective bacteria of the genus Pseudomonas produce sphingolipids that influence the worm's sphingolipid metabolism and thus support the host's protection against pathogens.
"This finding is relatively new," explains Peters, member of the CRC 1182. "Normally, bacteria use the sphingolipid metabolism of host organisms to manipulate it in a targeted manner to promote infections. In our case, however, we observe the opposite—bacterial sphingolipids apparently actively support the protection of the host."
Sphingolipids are fat-like molecules that are typically found in eukaryotes, where they fulfill important structural and regulatory functions, but are rare in bacteria. In Pseudomonas, they are synthesized via a previously unknown, alternative metabolic pathway—not as a component of primary metabolism, as is usually the case, but as a so-called secondary metabolite.
The researchers discovered that this previously unknown metabolic pathway is based on a specific biosynthetic gene cluster, a so-called polyketide synthase. "With our experiments, we were able to confirm that the worms survived better in the presence of Pseudomonas fluorescens bacteria possessing this gene cluster when they were infected with the pathogen Bacillus thuringiensis," emphasizes Peters, first author of the study.
After identifying the responsible genes, the scientists could confirm through further analyses that the gene cluster encodes the enzymes required for sphingolipid synthesis. "It is exciting to be authors on this important, breakthrough paper. We are pleased that our expertise in bacterial sphingolipid research has helped discover a new role in the worm microbiome for these enigmatic lipids," says Prof. Campopiano.
"The protective mechanism against infections with B. thuringiensis apparently works indirectly. The lipids produced by Pseudomonas influence the worm's sphingolipid metabolism, which presumably leads to an improved barrier function of the intestinal cells," explains Peters.
When the worm is infected with B. thuringiensis, the toxins of the pathogen create small pores in the cell membrane of the host, which makes it easier for the pathogens to penetrate. "We assume that the sphingolipid metabolism modified by P. fluorescens strengthens the stability and resistance of the cell membranes—and thus offers effective, indirect protection against pathogens," Peters continues.
"Overall, the new research work expands our understanding of how microbial metabolites support host defense against pathogens," says Dierking, independent group leader in the Evolutionary Ecology and Genetics research group.
In the long term, the researchers of the CRC 1182, who are also active in Kiel University's priority research area Kiel Life Science (KLS), hope that better knowledge of such fundamental mechanisms will also make it possible to influence disorders of the human gut microbiome, which may result in better treatment options for a variety of associated diseases.
More information: Lena Peters et al, Polyketide synthase-derived sphingolipids mediate microbiota protection against a bacterial pathogen in C. elegans, Nature Communications (2025).
Journal information: Nature Communications , Current Biology
Provided by Kiel University