With help from AI, researchers upgrade plants' internal alarm system to fend off pathogens

Sadie Harley
scientific editor

Andrew Zinin
lead editor

Scientists at the University of California, Davis, used artificial intelligence to help plants recognize a wider range of bacterial threats — which may lead to new ways to protect crops like tomatoes and potatoes from devastating diseases. The was published in Nature Plants.
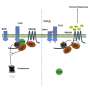
Plants, like animals, have immune systems. Part of their defense toolkit includes immune receptors, which give them the ability to detect bacteria and defend against it. One of those receptors, called FLS2, helps plants recognize flagellin — a protein in the tiny tails bacteria use to swim. But bacteria are sneaky and constantly evolving to avoid detection.
"Bacteria are in an arms race with their plant hosts, and they can change the underlying amino acids in flagellin to evade detection," said lead author Gitta Coaker, professor in the Department of Plant Pathology.
To help plants keep up, Coaker's team turned to using natural variation coupled with artificial intelligence — specifically AlphaFold, a tool developed to predict the 3D shape of proteins and reengineered FLS2, essentially upgrading its immune system to catch more intruders.
The team focused on receptors already known to recognize more bacteria, even if they weren't found in useful crop species. By comparing them with more narrowly focused receptors, the researchers were able to identify which amino acids to change.
"We were able to resurrect a defeated receptor, one where the pathogen has won, and enable the plant to have a chance to resist infection in a much more targeted and precise way," Coaker said.
Coaker said this opens the door to developing broad-spectrum disease resistance in crops using predictive design.
One of the researchers' targets is a major crop threat: Ralstonia solanacearum, the cause of bacterial wilt. Some strains of the soil-borne pathogen can infect more than 200 plant species, including staple crops like tomato and potato.
Looking ahead, the team is developing machine learning tools to predict which immune receptors are worth editing in the future. They're also trying to narrow down the number of amino acids that need to be changed.
This approach could be used to boost the perception capability of other immune receptors using a similar strategy.
Other authors of the study include Tianrun Li, Esteban Jarquin Bolaños, Danielle M. Stevens and Hanxu Sha of UC Davis and Daniil M. Prigozhin of Lawrence Berkeley National Laboratory.
More information: Tianrun Li et al, Unlocking expanded flagellin perception through rational receptor engineering, Nature Plants (2025).
Journal information: Nature Plants
Provided by UC Davis