Study uncovers how harmful RNA clumps form—and a way to dissolve them

Lisa Lock
scientific editor

Robert Egan
associate editor

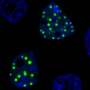
Look inside a brain cell with Huntington's disease or ALS and you are likely to find RNA clumped together. These solid-like clusters, thought to be irreversible, can act as sponges that soak up surrounding proteins key for brain health, contributing to neurological disorders.
How these harmful RNA clusters form in the first place has remained an open question.
Now, University at Buffalo researchers have not only uncovered that tiny droplets of protein and nucleic acids in cells contribute to the formation of RNA clusters but also demonstrated a way to prevent and disassemble the clusters.
Their findings, described in a study in Nature Chemistry, use an engineered strand of RNA known as an antisense oligonucleotide that can bind to RNA clusters and disperse them.
"It's fascinating to watch these clusters form over time inside dense, droplet-like mixtures of protein and RNA under the microscope. Just as striking, the clusters dissolve when antisense oligonucleotides pull the RNA aggregates apart," says the study's corresponding author, Priya Banerjee, Ph.D., associate professor in the Department of �鶹��Ժics, within the UB College of Arts and Sciences. "What's exciting about this discovery is that we not only figured out how these clusters form but also found a way to break them apart."
The study sheds new light on how RNA clusters form within biomolecular condensates.
Cells make these liquid-like droplets from RNA, DNA and proteins—or a combination of all three. Banerjee's team has researched them extensively, investigating their role in both cellular function and disease, as well as their fundamental material properties that present new opportunities for synthetic biology applications.
The condensates are essentially used as hosts by repeat RNAs, disease-linked RNA molecules with abnormally long strands of repeated sequences. At an early timepoint, the repeat RNAs remain fully mixed inside these condensates, but as the condensates age, the RNA molecules start clumping together, creating an RNA-rich solid core surrounded by an RNA-depleted fluid shell.
"Repeat RNAs are inherently sticky, but interestingly, they don't stick to each other just by themselves because they fold into stable 3D structures. They need the right environment to unfold and clump together, and the condensates provide that," says the study's first author, Tharun Selvam Mahendran, a Ph.D. student in Banerjee's lab.
"Crucially, we also found that the solid-like repeat RNA clusters persist even after the host condensate dissolves," Mahendran adds. "This persistence is partly why the clusters are thought to be irreversible."
The team was first able to demonstrate that repeat RNA clustering can be prevented by using an RNA-binding protein known as G3Bp1 that is present in cells.
"The RNA clusters come about from the RNA strands sticking together, but if you introduce another sticky element into the condensate, like G3BP1, then the interactions between the RNAs are frustrated and clusters stop forming," Banerjee says.
"It's like introducing a chemical inhibitor into a crystal-growing solution, the ordered structure can no longer form properly. You can think of the G3BP1 as an observant molecular chaperone that binds to the sticky RNA molecules and makes sure that RNAs don't stick to each other."
In order to reverse the clusters, the team employed an antisense oligonucleotide (ASO). Because ASO is a short RNA with a sequence complementary to the repeat RNA, it was able to not only bind to the aggregation-prone RNAs but also disassemble the RNA clusters.
The team found that ASO's disassembly abilities were highly tied to its specific sequence. Scramble the sequence in any way, and the ASO would fail to prevent clustering, let alone disassemble the clusters.
"This suggests our ASO can be tailored to only target specific repeat RNAs, which is a good sign for its viability as a potential therapeutic application," Banerjee says.
Banerjee is also exploring RNA's role in the origin of life. He is studying whether biomolecular condensates may have protected RNA's functions as biomolecular catalysts in the harsh prebiotic world.
"It really just shows how RNAs may have evolved to take these different forms of matter, some of which are extremely useful for biological functions and perhaps even life itself—and others that can bring about disease," Banerjee says.
More information: Tharun Selvam Mahendran et al, Homotypic RNA clustering accompanies a liquid-to-solid transition inside the core of multi-component biomolecular condensates, Nature Chemistry (2025).
Journal information: Nature Chemistry
Provided by University at Buffalo