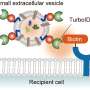
Graphical abstract. Credit: Journal of the American Chemical Society (2025). DOI: 10.1021/jacs.5c07631
Certain types of biochemical processes can impair the immune system's ability to recognize and kill cancer cells. Purdue University's W. Andy Tao and his associates have developed a new way to study these processes. They demonstrated the validity of their method in experiments involving leukemia and rare liver cancer cell lines.
Tao and 10 co-authors of their new method in the Journal of the American Chemical Society. Their work provides a system for tracking and identifying the various types of proteins and an unheralded but widely secreted class of bioparticles called extracellular vesicles (EVs) that can compromise immunotherapy.
EVs deliver a wide variety of cargo from one cell to another, including ribonucleic acid (RNA), a biomolecule that plays key roles in cellular and viral processes like protein formation. The new method relies on both RNA-binding proteins and EVs, which biomedical researchers have linked to many types of cancer under abnormal conditions.
"RNA-binding proteins are important since proteins typically do major work for the function of cells," said Tao, professor of biochemistry and a member of the Purdue Institute for Cancer Research. In recent years, new roles for RNA have been discovered, a trend that is driving many efforts to develop RNA-based tools, profile RNA-binding proteins and determine their functions.
The new method helps identify what kind of RNA-interacting proteins carried by EVs affect immune cells. Tao has an interest in EVs derived from tumors, which deliver RNAs that can influence immune responses while interacting with RNA-binding proteins in recipient cells.
"However, systematic profiling of these interactions remains limited largely due to the low-throughput nature of current methods," Tao and his co-authors reported. The strategies now available for profiling RNA-binding proteins on a large scale were not designed to selectively detect proteins derived from EVs that interact with RNA in recipient cells.
The new method addresses that issue. The method labels the RNA with a synthetic organic molecule that responds to ultraviolet light. Exposure to UV light cross-links (chemically connects) any nearby proteins.
"Through the EVs, these labeled RNAs will be delivered into the immune cells. Then we use UV to crosslink the proteins in the recipient cell," Tao explained. Isotopically labeling the proteins that come from the recipient cells helps scientists to tell them apart from those contained in the original cells.
Data science requires high-throughput experiments, Tao noted. But along with high throughput comes the need to ensure a low false-discovery rate. "Otherwise, garbage in, garbage out," he said. "For this high-throughput profiling experiment, we would like to make sure we have a low false-discovery rate. That's the part we try to control by involving these two labeling steps."
Tao's team validated the method's effectiveness in experiments tracking the interactions between EV RNA-binding proteins and Jurkat T cells, a cell line that researchers widely use to study leukemia. Additional tests similarly verified the method's ability to identify large numbers of EV RNA-binding proteins in immune cell lines infected with human intrahepatic cholangiocarcinoma. These cells, derived from a rare type of liver cancer, carry a mutation of biomedical interest because they resist immunotherapy, he said.
Immunotherapy relies on the immune system to recognize and kill cancer cells. Checkpoint proteins play a key role in this process, but their ability to recognize cancer cells can become compromised. EVs carry checkpoint proteins into immune cells, just as they do with RNA-binding proteins. Research in Tao's lab and elsewhere continues to explore what role EVs may play in similarly compromising immune cells.
"The role of the EV is certainly being recognized more and more," Tao said.
More information: Zheng Zhang et al, Proteomic Tracking Extracellular Vesicle RNA Interactors in Recipient Immune Cells through Orthogonal Labelings, Journal of the American Chemical Society (2025).
Journal information: Journal of the American Chemical Society
Provided by Purdue University