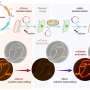
Inspired by bacteria's defense strategies, scientists develop chemical DNA tagging for genome editing

Gaby Clark
scientific editor

Robert Egan
associate editor

A research team led by scientists from the Helmholtz Institute for RNA-based Infection Research (HIRI) has introduced a new way to fine-tune genetic material. Their study, in Nature Biotechnology, describes an innovative technique in which chemical tags are attached directly to DNA, opening the door to new approaches in medicine, agriculture, and biotechnology.
Targeted editing of genetic information has advanced at an extraordinary pace in recent years. Tools such as the CRISPR-Cas9 "gene scissors" and base editing—a technique that makes precise, single-letter changes to DNA without cutting it—have already become standard in research and clinical development. These technologies are being used to treat genetic disorders, enhance crop resistance, and engineer bacteria for biotechnological purposes.
Researchers at the HIRI, a site of the Braunschweig Helmholtz Center for Infection Research (HZI), in cooperation with the Julius-Maximilians-Universität Würzburg (JMU), have developed a new method for precisely editing DNA. The HIRI team also cooperated with North Carolina State University in the U.S. and ETH Zurich in Switzerland. Their aim was to make genetic changes in bacteria, plants, and human cells even more accurate and gentle.
The team took inspiration from a natural bacterial defense system against viruses known as bacteriophages. To fight off these invaders, bacteria use two enzymes, DarT2 and DarG. During a viral infection, DarT2 attaches a chemical marker to the DNA, blocking replication and halting viral spread.
In the absence of a threat, DarG shuts down DarT2 and actively removes the marker. This finely tuned mechanism helps prevent the virus from spreading—and now serves as the foundation for a new genome editing approach.
This newly developed form of attachment has been named "append editing" by the researchers. "For the first time, this allows us to achieve new types of genetic modifications not possible with previous methods," the scientists explain.
To understand the mechanism, DNA can be imagined as a notebook in which each page consists of a long chain of letters. While traditional gene-editing techniques typically remove or replace individual letters within this chain, append editing introduces a small chemical group—ADP-ribose molecules—at a specific site.
This addition functions like a "sticky note" affixed to a particular letter. The chemical marker convinces the cell to change this DNA with high precision and minimal disruption. The type of change, however, depended on the organism in which it was introduced.
'DarT2'—pioneering a new era of genome editing
Unlike previous technologies, where the same tools produce similar results across all organisms, the effects of the append editing method were different between bacteria and eukaryotes, such as fungi, plants, and human cells.
"We observed that append editing led to the incorporation of large edits in bacteria based on a provided template, while in eukaryotic cells, the modified DNA base changed identity," explains Chase Beisel, affiliated department head at HIRI.
"This was one of the most surprising findings—that the outcome of DNA repair could be very different between organisms," adds Constantinos Patinios, a former postdoc in Beisel's lab.
Researchers see numerous potential applications for this tool. "Our append editing method greatly expands the toolkit of genome research and opens new doors for precision biotechnology and medical therapy development," says Darshana Gupta, a doctoral student at HIRI.
Specifically, microbes could be modified in a targeted manner—for example, to optimize naturally beneficial bacteria in the human body or to study pathogens more effectively. In human cells, precise editing could one day help to gently correct inherited diseases and provide new insights into DNA repair processes.
Further research is still needed before such applications can reach clinical practice. However, the scientists are confident. "DarT2 is another great example of the use of bacterial defenses in genome research," says Harris Bassett, who is completing his Ph.D. in Beisel's lab.
More information: Darshana Gupta et al, Targeted DNA ADP-ribosylation triggers templated repair in bacteria and base mutagenesis in eukaryotes, Nature Biotechnology (2025).
Journal information: Nature Biotechnology
Provided by Helmholtz Centre for Infection Research