New technology illuminates protein interactions in living cells
While fluorescence has long been used to tag biological molecules, a new technology developed at Yale allows researchers to use tiny fluorescent probes to rapidly detect and identify protein interactions within living cells while avoiding the biological disruption of existing methods, according to a report in Nature Chemical Biology.
Proteins are commonly tagged using variants of the “green fluorescent protein” (GFP), but these proteins are very large and are often toxic to live cells. They also tend to aggregate, making them difficult to work with and monitor. This new methodology uses the fluorescence emitted by a small molecule, rather than a large protein. It gives researchers a less disruptive way to capture images of the intricate contacts between folded regions of an individual protein or the partnerships between proteins in a live cell.
“Our approach bypasses many of the problems associated with fluorescent proteins, so that we can image protein interactions in living cells,” said senior author Alanna Schepartz, the Milton Harris Professor of Chemistry, and Howard Hughes Medical Institute Professor at Yale. “Using these molecules we can differentiate alternative or misfolded proteins from those that are folded correctly and also detect protein partnerships in live cells.”
Each protein is a three-dimensional structure created by “folding” its linear chain of amino acids. Usually only one shape “works” for each protein. The particular shape a protein takes depends on its amino acids and on other processes within the cell.
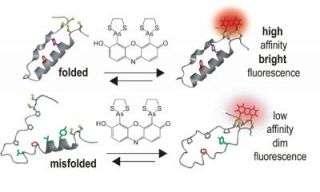
Schepartz and her team devised their new tagging system using small molecules, called “profluorescent” biarsenal dyes. These molecules easily enter cells and become fluorescent when they bind to a specific amino acid tag sequence within a protein. While these compounds have been used for about a decade to bind single proteins, this is the first time they have been used to identify interactions between proteins.
The researchers’ strategy was to split the amino acid tag for the dye into two pieces, locating each piece of the tag far apart in the chain of a protein they genetically engineered and expressed in the cells. Then they monitored cells exposed to the dye. Where the protein folded correctly, the two parts of the tag came together and the fluorescent compound bound and lit up. There was no signal unless the protein folded normally.
“This method of detection can provide important insights into how proteins choose their partners within the cell — choices that may be very different from those made in a test tube,” said Schepartz. She emphasizes that this technology does not monitor the process of protein folding — but, rather “sees” the protein conformations that exist at a given time.
“In theory, our technique could be used to target and selectively inactivate specific protein complexes in the cell, as therapy, or to visualize conformations at very high resolution for diagnostic purposes,” said Schepartz. She speculates that the technology could be applied to detection strategies that identify protein misfolding in neurodegenerative diseases like Alzheimer’s or Parkinson’s.
Source: Yale University

















