Scientists shrink the genetic code of E. coli to contain only 57 of its usual 64 codons

Krystal Kasal
contributing writer

Gaby Clark
scientific editor

Robert Egan
associate editor

The DNA of nearly all life on Earth contains many redundancies, and scientists have long wondered whether these redundancies served a purpose or if they were just leftovers from evolutionary processes. Both DNA and RNA contain codons, which are sequences of three nucleotides that either provide information about how to form a protein with a specific amino acid or tell the cell to stop (a stop signal) during protein synthesis.
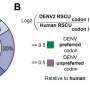
Altogether, there are 64 possible codon combinations and these combinations are nearly universal for all life on Earth. But some codons are redundant. There are only 20 amino acids available for a cell to work with, and 61 of the 64 codons are available for protein synthesis, while 3 are used as stop signals. This makes for a lot of redundancy in codons.
Some studies suggest that these redundancies might help prevent mutations in DNA, but reducing the genetic code of certain organisms by removing unnecessary parts can also be beneficial. In 2019, a group of scientists reduced the genome of E. coli to 61 codons from 64 by making 18,214 changes. They called the resulting version Syn61 and this virus-resistant version is being used to create more reliable drugs and for manufacturing novel materials.
Now, another group of scientists, some of whom worked on Syn61, have managed to further reduce the genetic code of E. coli down to 57 codons, making Syn57. They recently their work in Science.
This further reduction was a massive effort to take on. The team made over 101,000 codon changes by dividing up the genome into 38 sections and meticulously swapping out redundant codons with synonymous codons—those that perform the same function. Each time a swap was made, the researchers had to determine if the swap would be detrimental to the viability of the bacteria before moving on.
They explain, "Mapping and fixing at each stage of the synthesis was often crucial to enabling the next step of the synthesis. These experiments provide a paradigm for integrating 'just in time' defect mapping and fixing of initial designs into synthetic schemes, such that local defects are identified and fixed early in the synthesis and longer range, potentially epistatic or synthetic lethal, defects are identified and fixed as they emerge in the assembly process."
In the end, the team successfully shortened the genetic code to 57 codons by replacing six sense codons and a stop codon with synonymous codons. The resulting bacteria made with the new code were indeed a living organism, but the researchers found that they grow around four times slower than the parent strain—a problem they hope to eventually fix. However, the new strain shows a distinct gene expression profile, which indicates broad physiological adaptation.
Some possible applications of this new strain include virus-resistant organisms for biotechnology and industry, and the synthesis of proteins and polymers with new properties. Overall, the researchers are optimistic about the potential for this new strain. Their work also raises questions about whether there are limits to reducing the number of codons or creating organisms with entirely novel biochemistries.
The study authors say, "This work exemplifies how genome synthesis can move the genome sequences of organisms into new regions of sequence space that may not have been accessed by natural life."
Written for you by our author , edited by , and fact-checked and reviewed by —this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a (especially monthly). You'll get an ad-free account as a thank-you.
More information: Wesley E. Robertson et al, Escherichia coli with a 57-codon genetic code, Science (2025).
Journal information: Science
© 2025 Science X Network