'Fingerprints' match simulations with reality
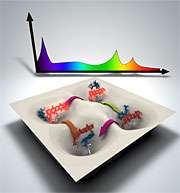
A theoretical technique developed at DOE's Oak Ridge National Laboratory is bringing supercomputer simulations and experimental results closer together by identifying common "fingerprints."
ORNL's Jeremy Smith collaborated on devising a method — dynamical fingerprints — that reconciles the different signals between experiments and computer simulations to strengthen analyses of molecules in motion.
"Experiments tend to produce relatively simple and smooth-looking signals, as they only 'see' a molecule's motions at low resolution," said Smith, who directs ORNL's Center for Molecular Biophysics and holds a Governor's Chair at the University of Tennessee. "In contrast, data from a supercomputer simulation are complex and difficult to analyze, as the atoms move around in the simulation in a multitude of jumps, wiggles and jiggles. How to reconcile these different views of the same phenomenon has been a long-standing problem."
The new method solves the problem by calculating peaks within the simulated and experimental data, creating distinct "dynamical fingerprints." The technique can then link the two datasets.
Provided by Oak Ridge National Laboratory