Scientists discover genetic markers for predicting seed oil quality
Researchers from Skoltech and Pustovoit All-Russian Research Institute of Oil Crops (VNIIMK) performed a genetic analysis of Russian sunflower lines and identified genetic markers that can help to predict the composition of tocopherols, one of the key attributes of oil quality. The research was published in G3: Genes, Genomes, Genetics.
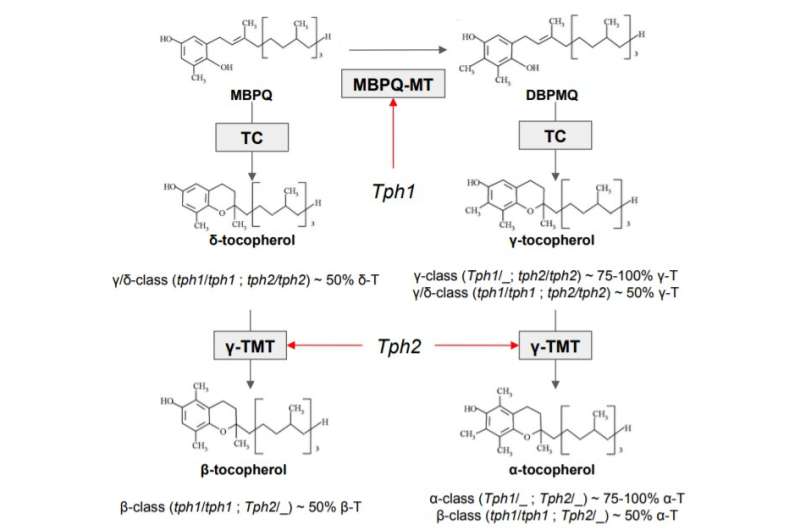
Tocopherols are a class of chemical compounds, many of which have Vitamin E activity. There are four types of tocopherols: alpha, beta, gamma, and delta. Vitamin E activity decreases, whereas antioxidant characteristics increase from alpha to delta. Dressing oils are produced using sunflower varieties with a high content of alpha- and beta-tocopherols that increase Vitamin E intake. By contrast, oils used for frying, baking and roasting require a higher content of gamma (and delta) tocopherols that reduce the form of thermal oxidation products during cooking.
Marker-assisted breeding that helps create new varieties based on genetic information is widely applied across the world. Researchers use DNA sequencing and large-scale genotyping to obtain genetic profiles of cultivated plants. Analyzing and comparing genetic profiles to field data can help to find genetic markers of traits useful for farming and use them to predict a plant's properties and value based on its genetic profile alone.
"We analyzed valuable breeding lines obtained by our colleagues from VNIIMK. To do this, we used high-throughput progeny genotyping for sunflower lines contrasting in tocopherol composition. In our genetic analysis, we tried to find out which parts of the plant's genome are related to tocopherol composition and discovered four genetic markers that allow predicting the composition of tocopherols in sunflower," Skoltech Ph.D. student Rim Gubaev, first author and co-founder of the OilGene startup, explains.
The identified genetic markers will help predict the composition of tocopherols for future sunflower lines and facilitate faster breeding of new varieties suitable for dressing and cooking oils production.
"The reason we chose sunflower is that it is a key source of vegetable oil, and Russia is the world's leading supplier of sunflower oil. The OilGene startup founded by Skoltech will use the markers to develop new testing tools," Skoltech researcher Stepan Boldrev, co-author and co-founder of OilGene, comments.
"Thanks to this project, we have gained valuable insights and built a team of like-minded people keen on helping breeders to introduce genetics in their work in order to create new commercial varieties. Our OilGene startup will focus on practical tasks and provide a genomic breeding service," Alina Chernova, Skoltech Ph.D. graduate and co-founder of OilGene, adds.
More information: Rim Gubaev et al, Genetic mapping of loci involved in oil tocopherol composition control in Russian sunflower (Helianthus annuus L.) lines, G3 Genes|Genomes|Genetics (2022).
Provided by Skolkovo Institute of Science and Technology


















