Molecular module coordinates plant cell wall formation and adaptive growth
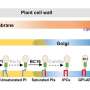
The plant cell walls represent the cellular basis for plant architecture and constitute the major component of plant biomass. Formation of multiple agronomic traits, e.g., plant height and mechanical force, largely depends upon orderly deposition of plant cell walls and precise control of cell wall biosynthesis. Therefore, cell wall accumulation is tightly coupled with various biological processes of plant growth and development.
Precise modulation of cell wall biosynthesis is required for plants given their exposure to fluctuating conditions and various stresses. Cell wall formation not only acts as the terminal output of multiple signaling pathways, but also is a central hub for integrating internal and external stimuli, which can balance various biological programs and facilitate the trade-off of multiple agronomic traits. Unraveling the molecular network of cell wall formation has been one of the hot topics in plant sciences, even though precise regulators that can sense signals and coordinate different biological processes are rarely reported, which impedes the development for rational design of agronomic traits in crop breeding.
In a study published in Molecular Plant, Prof. Zhou Yihua's group from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences reported that the receptor-like cytoplasmic kinase (RLCK)-VND6 module, a fine-tuning signaling cascade, coordinates cell wall accumulation and growth plasticity in rice.
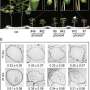
Through transcriptome deep sequencing and genome-wide co-expression assays, an unknown function RLCK that is co-expressed with secondary cell wall Cellulose Synthases was identified (named as CSK1) for in-depth characterization. CSK1 expression can be induced by multiple stress treatments, indicating it as an integrating pivot of various stress signals. The increases in cellulose content and wall thickness of vessels and fiber cells observed in the csk1 mutants, as well as the more vigorous xylem transport capacity of the mutants, define CSK1 a negative regulator for cell wall formation.
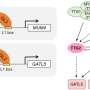
CSK1 is a nucleus- and cytoplasm-localized protein with bona fide kinase activity. An RNA-seq analysis from wild type and the csk1 mutant revealed broad differentially expressed genes enriched in GO terms related to cell wall, suggesting that CSK1 might help control the secondary cell wall regulatory network. The master regulator VND6 for secondary cell wall formation was then identified as an interacting and phosphorylating substrate of CSK1. VND6 was found positively regulating secondary cell wall biosynthesis according to the reductions in cellulose content and wall thickness of vessels and fiber cells in the vnd6 mutants. While VND6 is phosphorylated by CSK1, its activation activity on the downstream gene MYB61 is attenuated, thereby alleviating the transcription of a suite of SCW-related genes and accumulation of cell walls.
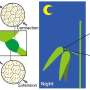
Therefore, CSK1-VND6 module acts as a "brake device" to precisely regulate the formation of secondary cell walls in rice. It was further found that CSK1 and VND6 implicate in abscisic acid-mediated stress signaling, which coordinates cell growth and SCW deposition.
This study identifies a molecular module for the precise control of cell wall biosynthesis and elucidates the molecular mechanism that coordinates secondary cell wall formation and adaptive growth, providing a tool for synergistic improvement of multiple traits for crop breeding.
More information: Shaoxue Cao et al, The RLCK-VND6 module coordinates secondary cell wall formation and adaptive growth in rice, Molecular Plant (2023).
Journal information: Molecular Plant
Provided by Chinese Academy of Sciences