Researchers uncover new mechanisms regulating plant response to temperature
Have you ever wondered why you get tired when the sun goes down? Why some flower petals open during the day and close at night? Or even how monarch butterflies know when to migrate south? Life on Earth has evolved to predict what time it is. This mechanism is referred to as the circadian clock: plants and animals alike have rhythmic, biological responses to the Earth's 24-hour and 365-day cycles using external cues like light and temperature.
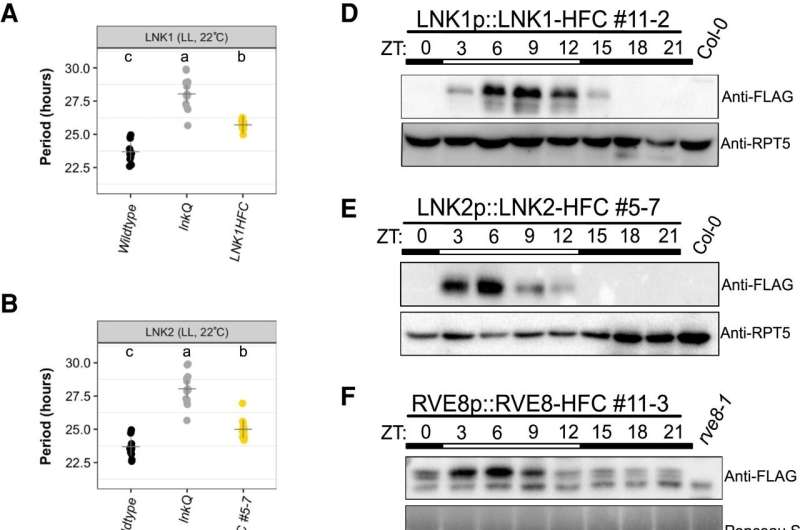
Research led by Dmitri Nusinow, Ph.D., associate member, Danforth Plant Science Center, and former Nusinow graduate student Maria Sorkin, Ph.D., has identified a new protein complex in plants that regulates temperature response by the circadian clock. As climate change influences daily and seasonal temperature patterns鈥攕uch as warmer nights and winters鈥攊t is critical to better understand how plants interpret and react to thermal cues.
Their findings, "COLD-REGULATED GENES 27 and 28 antagonize the transcriptional activity of the RVE8/LNK1/LNK2 circadian complex," were recently published in the journal Plant 麻豆淫院iology.
"The clock is essential for plants to correctly respond to temperature stimuli," wrote first author Sorkin, and scientists have uncovered a variety of ways in which the circadian clock helps plants acclimate to changes in temperature and survive stress鈥攅specially in model species like Arabidopsis.
"The circadian clock in Arabidopsis is well-studied," mentioned Sorkin, "so the most exciting part of this project was finding a brand-new protein complex that regulates temperature responses. No one else had discovered this interaction, even in an established system."
The complex comprises three proteins that interact in the evening to adjust to cooler temperatures. The research team importantly identified the mechanistic connection between these proteins and the specific time of days at which their interactions occur.
"Sorkin went to heroic lengths to discover how these three protein 'puzzle pieces' come together," said Nusinow. "We are always looking for protein complexes in our work, but we don't know how they will interact. Maria's dedication solved that puzzle," he continued.
Their findings are the result of three years of hard work鈥攕ometimes at odd hours late at night and early in the morning鈥攖o demystify how and when these proteins work together. Interestingly, the team "saw new complexes formed when we ran our experiments at different times of the day," Nusinow commented, "even just hours apart from each other."
The researcher's experiments involved collaboration with the Danforth Center's Proteomics & Mass Spectrometry Facility (PMSF) and plant growth team. The PMSF deployed state-of-the-art instrumentation to identify hundreds of potential proteins for the team to explore. In addition, collaborators from the University of Freiburg in Germany, the Plant-Environment Signaling Group at Utrecht University, and the Fundaci贸n Instituto Leloir, Instituto de Investigaciones Bioqu铆micas de Buenos Aires鈥揅onsejo Nacional de Investigaciones Cient铆ficas y T茅cnicas in Argentina, generously shared plant material for analysis of these proteins.
The Nusinow lab is excited to continue to study this protein complex at different temperatures with Stefanie King, a co-author and second-year graduate student at Washington University in St. Louis. "I'm grateful to learn from Maria and design experiments to look at the structure and regulation of the complex as a whole," King said.
Now that the researchers demonstrated that the protein complex interacts at specific times of day, they are interested in further understanding the interaction under varying temperatures. In addition, Stefanie is looking forward to mentoring an NSF REU intern in these techniques over the summer.
More information: Maria L Sorkin et al, COLD REGULATED GENE 27 and 28 antagonize the transcriptional activity of the RVE8/LNK1/LNK2 circadian complex, Plant 麻豆淫院iology (2023).
Provided by Donald Danforth Plant Science Center



















