A blueprint for making cereal crops more resistant to fungal disease

Powdery mildew is a destructive fungal disease of barley that can result in crop losses of up to 40%. To protect themselves from powdery mildew, barley has evolved a series of immune receptors that each recognize matching, strain-specific powdery mildew proteins known as effectors.
This recognition event confers resistance and so insights gleaned from a study of this interaction could be exploited by scientists to make barley and its sister species such as wheat more resistant to this costly disease.
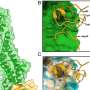
Now, using cutting-edge technology, scientists from the Max Planck Institute for Plant Breeding Research (MPIPZ) in Cologne, Germany, have succeeded in determining the structure formed by one immune receptor, called MLA13, in complex with its matching fungal effector, AVRA13-1. Their findings are in the EMBO Journal.
The researchers, led by Paul Schulze-Lefert at the MPIPZ, Elmar Behrmann at the University of Cologne and Jijie Chai at Westlake University in Hangzhou, China, used the cryogenic electron microscopy (cryo-EM) technique. In cryo-EM, samples are cooled to cryogenic temperatures and the structures of biological specimens such as proteins are preserved by embedding in an amorphous form of ice.
The resulting structure with atomic resolution reveals how the plant immune receptor and fungal effector interact with each other as well as the structure adopted by the fungal effector.
These insights allowed first author Aaron W. Lawson to engineer a new version of another immune receptor, MLA7, which recognizes an effector called AVRA7. The sequences of MLA immune receptors are highly similar to each other, which is also the case for MLA7 and MLA13.
Lawson and his co-authors thus asked if, based on the MLA13-AVRA13-1 structure, they could change the recognition specificity of MLA7. Indeed, by changing only one amino acid in the protein sequence of MLA7, the authors succeeded in engineering a new version of MLA7 that now recognizes AVRA13-1, while retaining its recognition of AVRA7.
Plant breeding traditionally involves painstaking and time-intensive crosses in order to obtain plants with the desired combination of different attributes. However, the fungus that causes powdery mildew on barley diversifies very rapidly, meaning that traditional breeding techniques cannot keep up with the emergence of new, virulent fungal variants.
In showing how immune receptors can be engineered to change or expand their specificity—a method that is much more precise and rapid than traditional breeding—the authors' findings show how structure-guided gene editing of such receptors could be a viable tool for protecting barley from disease and ensuring food security.
Genes encoding MLA immune receptors evolved in a common ancestor of a grass family that includes the sister species barley, wheat, oats and rye, and are found in each of these cereals. Since MLA immune receptors can also confer immunity to other microbial pathogens such as rust fungi and the rice blast fungus, gene-edited MLA receptors have the potential to protect these staple crops from multiple economically relevant diseases.
More information: Aaron W Lawson et al, The barley MLA13-AVRA13 heterodimer reveals principles for immunoreceptor recognition of RNase-like powdery mildew effectors, The EMBO Journal (2025).
Journal information: EMBO Journal
Provided by Max Planck Society