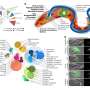
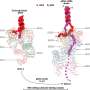
Cryo-EM structure of the decorated DMT from T. brucei. Credit: Science (2025). DOI: 10.1126/science.adr3314
Millions of people worldwide are affected by African sleeping sickness, Chagas disease and other life-threatening infections caused by microscopic parasites borne by insects such as the tsetse fly.
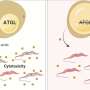
Each of the underlying single-celled parasites—Trypanosoma brucei and its relatives—has one flagellum, a whiplike appendage that is essential for moving, infecting hosts and surviving in different environments.
Now, a research team at the California NanoSystems Institute at UCLA, or CNSI, has applied leading-edge atomic imaging and AI-driven modeling to create the most detailed 3D map yet of the flagellum on Trypanosoma brucei, which causes sleeping sickness. The study, in the journal Science, identified 154 different proteins that make up the flagellum, including 40 that are unique to the parasite.
By capturing the molecular motors that drive the parasite's movement during a key transitional state, the investigators developed a new model for how they swim through blood and tissue. The findings shed light on a critical mechanism essential to Trypanosoma brucei's survival, transmission to hosts and disease processes. This detailed view of the parasite's flagella could help drive progress in treating the illness they cause.
"Our study provides a complete molecular blueprint of the flagellum's structural framework, explaining how its movement is powered at an atomic level," said co-corresponding author Z. Hong Zhou, a professor of microbiology, immunology and molecular genetics at the UCLA College and founding director of CNSI's Electron Imaging Center for Nanosystems, or EICN.
"By leveraging AI-driven structural modeling, we uncovered unique parasite-specific proteins that contribute to flagellar architecture and function."
A three-dimensional map of the basic structural unit in the parasite Trypanosoma brucei’s flagellum, with various mechanical and motor proteins labeled. Credit: California NanoSystems Institute at UCLA
How the parasite was mapped using the cryoEM
The imaging technique used in the study was cryogenic-electron microscopy, or cryoEM, in which frozen biological samples are probed with electrons to reveal details impossible to capture with visible light. Maps generated with cryoEM received further analysis using artificial intelligence tools, such as an algorithm for predicting a protein's shape based on the amino acids that make it up.
The scientists found that tiny motor-like structures in the microbe's flagellum create motion by acting in a coordinated fashion, similar to the way rowers in a dragon boat synchronize their strokes to move through water.
"Trypanosomes have evolved specialized motion to survive in both the tsetse fly and the human bloodstream, making their flagellum a central feature of their biology," said co-corresponding author Kent Hill, a UCLA professor of microbiology, immunology and molecular genetics and a CNSI member. "By understanding how their unique structural features contribute to movement, we gain insight into fundamental aspects of parasite adaptation and host interactions."
Potential future implications
Sleeping sickness initially manifests as fever, headaches, joint pain and itching. After the parasite reaches the central nervous system, the disease can progress to spur severe neurological symptoms.
The study may provide potential targets for therapies that effectively eliminate the parasite or block its transmission to humans, as well as give clues about how to address illnesses caused by other related microbes.
Beyond medical treatment, the insights into an understudied microbe could have impacts such as elucidating details of earlier stages in evolution and inspiring engineers who borrow from nature to inform their designs.
More information: Xian Xia et al, Trypanosome doublet microtubule structures reveal flagellum assembly and motility mechanisms, Science (2025).
Provided by University of California, Los Angeles