Innovative bead design will enhance single-cell transcriptomics accuracy

Single-cell transcriptomics has revolutionized the study of cellular diversity and function by enabling gene expression analysis at single-cell resolution. The technique is crucial for understanding how different cells function, how they communicate, and how they contribute to various biological processes and diseases.
However, technical errors in the synthesis of oligonucleotides (short DNA sequences)—particularly in droplet-based methods such as Drop-seq and 10x Chromium—frequently lead to inaccurate results, causing researchers to discard valuable data. These synthesis errors obscure identification of the cell barcode (which identifies the cell) and UMI (unique molecular identifier) resulting in substantial data loss and limiting sequencing efficiency.
In an article in Communications Biology, Associate Professor Adam Cribbs and Dr. Jianfeng Sun at the Botnar Institute for Musculoskeletal Sciences identified a major source of error: premature truncation of UMIs before sequencing. To address this, they developed a novel bead design that improves the reliability of gene expression measurements by mitigating truncation errors in UMIs prior to sequencing.
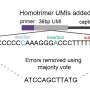
They added what they call an "anchor" between two important parts of the sequence: between the barcode and UMI. This anchor provides a clear demarcation point, enhancing UMI recognition and minimizing synthesis errors.
The work builds on the Oxford team's previous homotrimer UMI error correction strategy, which focused on errors arising after sequencing. In contrast, the new anchor-interposed approach tackles UMI errors at the pre-sequencing stage, further advancing long-read transcriptomics.
The researchers found that the new bead design enhances UMI identification, improving molecular quantification. UMIs are critical for counting the number of unique RNA molecules captured from each cell. They also observed a marked increase in the number of detected transcripts per cell, enhancing the reliability of single-cell sequencing.
In addition, the approach effectively counteracts synthesis limitations, such as oligonucleotide purification challenges, with only a small additional nucleotide sequence required.
Dr. Sun, first author of the study, explained, "By incorporating a four-base anchor sequence into droplet-based beads, we have improved barcode and UMI recognition, leading to a greater number of high-quality long reads. This new bead design enhances the overall reliability of single-cell sequencing."
Beyond its direct impact on sequencing accuracy, the study has broad implications for single-cell biology. Advancing the technical capabilities of single-cell sequencing is crucial for accelerating biological discovery and understanding complex cellular systems to advance science and improve health care outcomes.
More information: Jianfeng Sun et al, Enhancing single-cell transcriptomics using interposed anchor oligonucleotide sequences, Communications Biology (2025).
Journal information: Communications Biology
Provided by University of Oxford