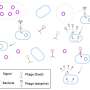
Examples of intracellular Phollow phage organization (a-b) Z-projected images of bacterial cells harboring viral foci. Images highlight two patterns of virion organization throughout the intracellular space: (a) a scattered distribution and (b) a serpentine pattern with the presence of ribbon structures. Images are pseudo-colored according to z-depth, representing a total of 1.36 μm. A white dashed line marks the cell perimeter. Credit: Nature Microbiology (2025). DOI: 10.1038/s41564-025-01981-1
Researchers at the University of California, Irvine have developed a live-imaging system, Phollow, that tracks individual bacteriophages as they spread through the gut of zebrafish, showing that phages from different bacterial hosts vary in how they replicate, disseminate, and interact with host tissues.
Bacteriophages, or phages, are viruses that infect and kill bacteria. They are the most numerous biological entities on Earth and among the earliest colonizers of the human gut. Phages play multiple roles in the human microbiome: they can influence microbial population dynamics, act as a form of innate immune modulation in newborns, and enhance bacterial fitness by mediating horizontal gene transfer.
Controlled phage outbreaks could potentially encourage the spread of beneficial activities, or be used to deplete resident microbes that become harmful under certain conditions. Transmission dynamics, the patterns of how phages replicate within one host, spread to others, and influence bacterial communities, would need to be comprehensively understood to use phage outbreaks productively.
Studying transmission dynamics is technically challenging, as the factors governing phage replication and transmission in situ are largely unknown. Without a contextualized understanding of location and duration of phage replication or whether phages are in the form of extracellular virions or intracellular prophages, progress in this area is severely constrained.
In the study, "Phollow reveals in situ phage transmission dynamics in the zebrafish gut microbiome at single-virion resolution," in Nature Microbiology, researchers designed a live-imaging approach to investigate how bacteriophages replicate and spread in bacterial communities and animal hosts.
Time-lapse movie depicting Phollow phage induction, assembly and dispersal. Time-series images of E. coli HS Phollow virocells growing on agar pads containing MMC. Optical frames were generated from four tile-scanned and merged fields of view, acquired at regular 10-min intervals over a period of 370 min. The time series starts at 0 min, where individual bacterial cells appear as small white rods. As time progresses, these cells undergo filamentation, a hallmark morphological change indicative of the SOS response induced by MMC genotoxicity. Subsequently, a subpopulation of bacterial cells displays the formation of intracellular fluorescent viral foci followed by explosive cell lysis. Upon lysis, Phollow phage virions can be seen dispersing throughout the extracellular milieu. Credit: Nature Microbiology (2025). DOI: 10.1038/s41564-025-01981-1
Phollow was applied in germ-free zebrafish colonized with engineered strains of E. coli and Plesiomonas, allowing visualization of phage behavior at single-virion resolution. Researchers constructed fluorescently tagged phages and introduced them into modified bacterial hosts referred to as Phollow virocells.
Viral replication was induced using antibiotics, and phage activity was monitored in vivo using time-lapse imaging, super-resolution microscopy, flow virometry, and expansion microscopy.
Phollow phages were observed to form viral aggregates that dispersed into clouds of rapidly diffusing particles following bacterial lysis. Peak induction of viral replication occurred one hour after treatment with mitomycin C and involved approximately 20% of the bacterial population.
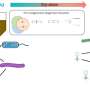
Design, construction and infectivity of Phollow phages. Credit: Nature Microbiology (2025). DOI: 10.1038/s41564-025-01981-1
At peak replication, Phollow virocells contained an average of 1.6 viral foci per micron of cell length. Three-dimensional projections revealed viral foci with surface areas approximately 100 times larger than a single P2-like phage capsid. Transmission electron microscopy found no overt structural differences between wild-type and Phollow phage virions.
Flow virometry showed that mitomycin C, ciprofloxacin, and trimethoprim all induced comparable peak virion output. Trimethoprim-induced outbreaks in zebrafish intestines produced clouds of phages throughout the gut within four hours, which largely disappeared by 24 hours. Infectious virions peaked at four hours in gut tissues and persisted in surrounding water over 24 hours. Plesiomonas-derived phages spread systemically to the liver and brain.
In vitro, Phollow phages were found replicating in new host cells and undergoing onward interbacterial transmission. In vivo, a second wave of phage replication confirmed horizontal transmission had occurred.
Initial testing of Phollow shows a capability to enable multiscale investigations of phage transmission and transkingdom interactions that may open new avenues for phage-based microbiome therapies.
Live imaging and flow virometry techniques can support experimental design aimed at dissecting the mechanics of phage outbreaks, with implications for a host of untapped clinical opportunities.
More information: Lizett Ortiz de Ora et al, Phollow reveals in situ phage transmission dynamics in the zebrafish gut microbiome at single-virion resolution, Nature Microbiology (2025).
Journal information: Nature Microbiology
© 2025 Science X Network