New mass-spectrometry technique boosts enzyme screening speed by up to 1,000 times

Scientists have developed a new technique to screen engineered enzyme reactions, which could lead to faster and more efficient creation of medicines and sustainable chemicals.
Enzymes are proteins that catalyze chemical reactions, turning one substance into another. In labs, scientists engineer these enzymes to perform specific tasks like the sustainable creation of medicines and materials.
These biocatalysts have many environmental benefits as they often produce higher product quality, lower manufacturing cost, and less waste and reduced energy consumption. But to find "the one," scientists must test hundreds of variants for their effectiveness, which is a slow, expensive, and resource-intensive process.
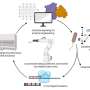
Research conducted by the University of Manchester in collaboration with AstraZeneca is changing this. The team developed a method for a technique that can test enzyme activity up to 1,000 times faster than traditional methods. The , developed over the last eight years and detailed today in the journal Nature Protocols, is called DiBT-MS (Direct Analysis of Biotransformations with Mass Spectrometry).
"Current screening methods can really slow things down when we're trying to find efficient biocatalysts because there are so many possible variations to test in an enzyme library. Some faster methods, like fluorescent microwell plates, do exist, but even then, you can hit a wall if your product doesn't naturally fluoresce. That's why there's a real need for a quicker, simpler way to screen reactions.
"Our research shows how DiBT-MS can dramatically speed up the process, skipping the need for complicated sample preparation and using significantly less solvent and sample material, ultimately lessening our impact on the environment," says Sabine Flitsch.
It builds on an existing technology called DESI-MS (Desorption Electrospray Ionization Mass Spectrometry), a powerful tool that allows scientists to analyze complex biological samples without the need for extensive sample preparation.
By making small adaptations to the technology, the scientists designed a protocol to directly analyze enzyme-triggered chemical reactions, known as biotransformations, in just minutes. The new method can process 96 samples in just two hours—tasks that would previously take days using older techniques.
It has also been optimized to allow the researchers to reuse sample slides multiple times, improving testing efficiency and decreasing the use of solvents and plasticware.
The team has already successfully applied this technique to a range of enzyme-driven reactions, including those enzymes particularly valuable in the development of therapeutics.
"This approach opens up enzyme research to a much wider range of laboratories. And as the demand for sustainable and cost-effective chemical production and higher throughput assay screening grows, DiBT-MS will be an essential tool.
"Its simplicity, high throughput, and minimal sample preparation makes it an ideal choice for biochemists and chemical biologists who need a reliable, efficient way to conduct rapid and comprehensive enzyme screening," says Perdita Barran.
Looking ahead, the University of Manchester will continue to explore ways to boost partnerships between laboratories and tackle other challenges that often hinder collaboration, such as geographical barriers and limited funding.
More information: Ruth Knox et al, Direct analysis of biotransformations with mass spectrometry—DiBT-MS, Nature Protocols (2025).
Journal information: Nature Protocols
Provided by University of Manchester