ExIGS bridges microscopy and sequencing to track nuclear abnormalities
Justin Jackson
褋ontributing writer

Sadie Harley
scientific editor

Robert Egan
associate editor

Harvard University researchers at the Broad Institute report that expansion in situ genome sequencing (ExIGS) linked nuclear abnormalities to hotspots of aberrant chromatin regulation, potentially eroding cell identity. The findings offer insight into age-related cell failure and showcase the ability of ExIGS to simultaneously sequence DNA and image proteins at nanoscale.
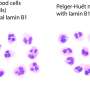
Under a microscope, cells can be identified by size, shape, structure, and the appearance of organelles. Morphology of the nucleus has proven especially important in pathology, as abnormalities in nuclear shape or chromatin texture are signs of mechanistic breakdowns, often informing cancer and blood disorder diagnoses.
Genomic sequencing has shifted focus toward epigenomic, transcriptomic, and proteomic measurements for defining cell types and states, allowing researchers a glimpse into the potential causes of disease morphology.
Microscopy observes the outcome of a failed mechanism, offering little information regarding causation. Sequencing can reveal the genomic underpinning of the change, but lacks tissue-specific context within the cell environment. Recent spatial genomics methods, such as ExIGS, have begun to integrate microscopy and sequencing to map transcriptomes within tissues at single-cell resolution.
In the study, "Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation," in Science, researchers developed ExIGS to sequence genomic DNA and localize nuclear proteins at nanoscale resolution within single cells.
ExIGS analyzed 63 healthy skin fibroblasts to measure read gains after expansion and 109 lung fibroblasts from a widely used cell line, all imaged for nuclear envelope and chromatin markers. Fibroblasts from a progeria patient provided 196 nuclei alongside 63 nuclei from healthy controls.
Progeria samples were chosen because progeria causes rapid aging of the nucleus via a defective lamin protein and disruptions in these cells are well characterized in pathology. Mouse heart tissue and fibroblasts from a 92-year-old donor broadened the aging comparison.
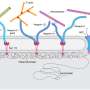
In Phase 1, samples underwent a two-step anchoring process to preserve both genomic DNA and protein signals within an expandable polymer matrix. In Phase 2, rolling circle amplification generated clonal nanoballs in the gel. The amplicon nanoballs are extracted and sequenced on a conventional Illumina platform. In Phase 3, multimodal data were integrated through a computational pipeline to capture what DNA was found at which location within the expanded cell.
Using ExIGS with expanded samples, more than 10 times as many DNA reads per nucleus were obtained while keeping the 3D layout intact. Approximately 33.6% of reads were mapped within 200 nm of nuclear proteins. DNA regions near the nuclear shell (lamin) tended to be inactive, whereas regions near nuclear speckles tended to be active.
In progeria patient cells, the nuclear shell thickened and folded inward, creating small areas where active DNA was forced close to the shell and appeared repressed. In normal, aged, and mouse heart cells, gene-reading enzymes were scarce within 200 nm of lamin folds, indicating that such folds shut down nearby gene activity.
The authors conclude that lamin organization acts as a key gatekeeper of gene activity, with abnormal lamin folds creating pockets of silenced DNA that may drive cell dysfunction in aging and disease. ExIGS offers a powerful new way to map how nuclear structure influences genome function, enabling studies of disorders like progeria, cardiovascular disease, and normal aging.
Written for you by our author , edited by , and fact-checked and reviewed by 鈥攖his article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a (especially monthly). You'll get an ad-free account as a thank-you.
More information: Ajay S. Labade et al, Expansion in situ genome sequencing links nuclear abnormalities to aberrant chromatin regulation, Science (2025).
Journal information: Science
漏 2025 Science X Network