Mapping corn DNA reveals how genes shape plant traits and pest resistance

Stephanie Baum
scientific editor

Andrew Zinin
lead editor

A team that includes Rutgers University-New Brunswick scientists has unlocked some of the secrets of corn DNA, revealing how specific sections of genetic material control vital traits such as plant architecture and pest resistance.
The discovery could enable scientists to use new technologies to improve corn, making it more resilient and productive, the scientists said.
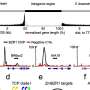
In a study in the science journal Nature Plants, researchers finding where certain proteins called transcription factors attach to the DNA in corn plants and how this sticking changes how genes are turned on or off in a particular tissue. They looked at two lines of corn and found big differences at these spots in the DNA sequence, which they said could help explain why the plants look and act differently.
"In this work, we discovered where transcription factors are binding in the genome and therefore influencing the expression of maize [corn] genes," said Andrea Gallavotti, a professor at the Waksman Institute of Microbiology and an author of the study. "Importantly, we did this analysis in two distinct maize [corn] lines that are different for many traits, including resistance to disease and architecture."
In North America, "corn" and "maize" refer to the same cereal grain. However, "maize" is the more internationally recognized and scientifically preferred term, said Gallavotti, also a professor in the Department of Plant Biology in the Rutgers School of Environmental and Biological Sciences.
Corn or maize touches many aspects of daily life worldwide. It is a staple food for many cultures around the world and is rich in carbohydrates, fiber, vitamins and minerals. It also has major industrial applications—used as livestock feed, for the production of biodegradable plastics, adhesives and textiles, and to produce ethanol.
The research is a collaborative effort between Rutgers and New York University scientists, led by Shao-shan Carol Huang, and other institutions, who are focused on tackling inquiries into the extremely complex and large maize genome. This partnership has been instrumental in advancing their understanding of what regulates when and where genes are turned on and off in maize, Gallavotti said.
The team started by looking to achieve a better understanding of how transcription factors modulate maize genes, adjusting, regulating or controlling their level of activity. Sifting through vast amounts of bioinformatic data, they created a map of the binding sites of transcription factors in the maize genome. The transcription factors affix themselves to special parts of the maize plant's DNA called cis-regulatory regions.
Once the researchers had this information, they were able to compare these binding sites across different maize lines to understand variations. The team contrasted two different types of maize plants, B73 and Mo17, in the study.
"We found that there are big differences in where transcription factors bind and in the organization of these cis-regulatory regions in the two types of maize," Gallavotti said. "These differences affect gene expression, and the resulting traits are an important source of variation in maize."
Using an extremely precise biological tool known as CRISPR-Cas9, the team edited some of these DNA regions and studied the effects of the changes on the plant, including on a gene regulating resistance to ear worms.
CRISPR stands for clustered regularly interspaced short palindromic repeats. It is a natural defense mechanism found in bacteria, used to protect themselves from viruses. Scientists have adapted this system for use in gene editing.
The system involves two key components. CRISPR RNA is a molecule that guides the system to the specific DNA sequence that needs to be edited. Cas9 is an enzyme or protein that acts like molecular scissors to cut the DNA at the targeted location.
"Variation in these cis-regulatory regions was crucial for the domestication and improvement of many crops," Gallavotti said. "Today, technologies like CRISPR-Cas9 allow us to introduce changes in certain traits, and cis-regulatory regions are important targets for these changes."
Until now, the challenge for scientists has been to figure out what to target.
"Our analysis helps map and study these regions, which can be used to improve crop species," Gallavotti said. "We hope this resource can be used to target particular regions for any trait. It could be resistance to stress, resistance to pests, modification of the architecture of a plant."
Rutgers researchers at the Waksman Institute of Microbiology who contributed to the study included Mary Galli, the main author of this study, Zongliang Chen, Amina Chaudhry, Jason Gregory and Fan Feng.
More information: Mary Galli et al, Transcription factor binding divergence drives transcriptional and phenotypic variation in maize, Nature Plants (2025).
Journal information: Nature Plants
Provided by Rutgers University