Discovery of a tRNA methyltransferase with an unusual domain architecture and functional features

Sadie Harley
scientific editor

Andrew Zinin
lead editor

All living organisms encode the amino acid sequences of proteins as nucleotide sequences (genetic information) in their genomic DNA. The genetic information is transcribed and subsequently translated into functional proteins.
Transfer RNA (tRNA) acts as an adaptor molecule that decodes codons on messenger RNA (mRNA) and delivers the corresponding amino acid to the ribosome, the machine of protein synthesis.
Following transcription, tRNAs undergo extensive chemical modifications, which play crucial roles in fine-tuning their structural stability and functional capacity.
Among the three domains of life, archaea are believed to have thrived in the harsh environments of early Earth, such as high temperature, high salinity, and low oxygen. To survive these extreme conditions, archaeal tRNA modifications are thought to have been diversified uniquely during evolution.
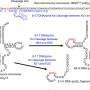
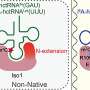
In 2019, the research group reported the nucleotide sequence of tRNATrp isolated from Thermococcus kodakarensis, a hyperthermophilic archaeon. This tRNA was found to contain a 2′-O-methylcytidine (Cm) modification at position 6. However, the methyltransferase responsible for this specific modification remained unknown.
In a new study, the researchers employed comparative genomics to examine the genome of T. kodakarensis along with those of closely related archaeal species, leading to the identification of candidate genes. Among these, the team focused on the gene TK1257. The study has been in Nucleic Acids Research.
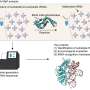
Through biochemical analysis using the purified recombinant protein and high-sensitivity RNA mass spectrometry, the researchers demonstrated that the TK1257 gene product catalyzes the formation of the Cm6 modification, identifying it as a previously uncharacterized tRNA methyltransferase.
Further sequence and structural modeling analyses revealed that the enzyme contains both a THUMP domain, which recognizes and binds the 3′-end of tRNA, and a SPOUT catalytic domain with a characteristic trefoil knot structure.
This domain architecture—combining THUMP and SPOUT domains—has not been reported previously in tRNA methyltransferases. Accordingly, the researchers named this enzyme TrmTS (Transfer RNA methylation gene product with THUMP and SPOUT domains).
TrmTS exhibits substrate specificity for adenosine, cytidine, and uridine, catalyzing 2′-O-methylation at the ribose moiety of these nucleosides. Notably, it does not modify guanosine, making its substrate selectivity highly unique among known tRNA methyltransferases.
Moreover, a T. kodakarensis mutant lacking the trmTS gene exhibited impaired growth under extreme heat conditions (93°C), suggesting that the Cm6 modification contributes to the thermal stability of tRNA and plays a role in adaptation to high-temperature environments.
This study presents a novel paradigm in tRNA modification biology, identifying a tRNA methyltransferase that does not conform to existing classification schemes.
The discovery of TrmTS, with THUMP and SPOUT domains, sheds new light on the diversity and evolution of tRNA modifications and provides important insights into the molecular strategies archaea use to thrive in extreme environments.
More information: Teppei Matsuda et al, A transfer RNA methyltransferase with an unusual domain composition catalyzes 2′-O-methylation at position 6 in tRNA, Nucleic Acids Research (2025).
Journal information: Nucleic Acids Research
Provided by Ehime University