3D holographic imaging tracks lysosomal changes in live cells without chemical labels

Sadie Harley
scientific editor

Robert Egan
associate editor

A team of researchers from the Institute of Applied Sciences and Intelligent Systems of the National Research Council of Italy (ISASI-CNR) and the Telethon Institute of Genetics and Medicine (TIGEM) has developed a method to observe lysosomes in live suspended cells—quantitatively, in 3D, and without the use of chemical labels.
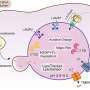
Their study, published in , showcases holographic tomography in flow cytometry configuration (HTFC) as a powerful tool for identifying morphological and spatial lysosomal changes in models of lysosomal storage diseases (LSDs), with a focus on Niemann-Pick type C1 (NPC1).
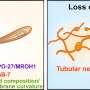
"This innovative approach could revolutionize the study of lysosomal storage disorders. For the first time, we can measure biophysical parameters of lysosomes—such as density and volume—and see how pathological accumulation of molecules alters their physical properties. These parameters can be used to study disease mechanisms, progression, and therapeutic response. In NPC1, we showed that correcting lysosomal positioning can resolve cholesterol accumulation," says Diego Medina, Principal Investigator at TIGEM.
A step closer to clinical application
Lysosomal storage diseases are a group of over 60 rare genetic disorders caused by enzyme or protein defects in lysosomes, leading to severe impacts on tissues and organs, particularly the central nervous system. Current diagnostic and monitoring methods are limited by a lack of tools that allow real-time functional analysis of lysosomes in live cells.
The HTFC technology provides label-free, high-resolution 3D imaging in a clinically relevant context—suspended live cells, instead of traditionally used adherent cells.
"For the first time, this technology allows us to obtain quantitative and three-dimensional information in suspended cells from Niemann-Pick type C models," explains Daniele Pirone, first author and researcher at ISASI-CNR. "This context is much closer to clinical reality."
Using HTFC, the researchers analyzed thousands of live cells in suspension, acquiring high-content 3D tomograms based on refractive index, without staining or complex preparation. They identified morphometric 3D biomarkers that can reliably distinguish healthy from diseased cells, and monitored the effects of pharmacological and genetic treatments.
"With this method, we can accurately measure changes in lysosomal morphology and positioning, paving the way for new biomarkers for LSDs," add Pasquale Memmolo, Senior Researcher at ISASI-CNR, and Lisa Miccio, Senior Researcher at ISASI-CNR.
From bench to bedside: Next steps
Looking forward, the team aims to validate the technology on patient-derived cells (e.g., fibroblasts and blood cells) and enhance spatial resolution to reach single-lysosome identification. These advancements would combine the benefits of high-resolution microscopy with large-scale statistical analysis.
"The integration of holographic cytometry into the translational research pipeline is a crucial step toward clinical application," concludes Pietro Ferraro, Research Director and Principal Investigator at ISASI-CNR.
"HTFC has tremendous potential as a diagnostic and therapeutic screening tool, and our results encourage us to move forward with patient cell validation."
More information: Daniele Pirone et al, Drug-Induced Reversible Lysosomal Changes Tracked in Live Cells by Holo-Tomographic Flow Cytometry, ACS Nano (2025).
Journal information: ACS Nano
Provided by Telethon Foundation