Bacteria reveal a complex arsenal of over 200 viral defense strategies

Stephanie Baum
scientific editor

Robert Egan
associate editor

Researchers at UT Southwestern Medical Center have identified more than 200 strategies used by bacteria to avoid viral infection. Their findings, in Cell Host & Microbe, shed light on a microbial "arms race" that could lead to new approaches to fighting infectious bacteria.
"There's a wealth of phage defense strategies in nature that we don't have a handle on," said Kevin Forsberg, Ph.D., Assistant Professor of Microbiology at UT Southwestern. "These findings could one day help improve therapies for treating antibiotic-resistant bacterial infections."
Dr. Forsberg co-led the study—the first independent publication of the Forsberg Lab—with Luis Rodriguez-Rodriguez, a fifth-year UTSW graduate student.
Like all living things, bacteria are under constant risk of infection by viruses. Those that infect bacteria are collectively known as bacteriophages, or "phages." Consequently, bacteria have evolved various ways to fight this threat, while viruses have developed strategies to evade bacterial defenses—the majority of which remain uncharacterized, Dr. Forsberg explained.
Most research to identify antiphage defenses has relied on the propensity of defense-related genes to cluster in the bacterial genome; thus, identifying one defense gene often leads to others, simply based on its location within each bacterium's single, circular chromosome. Although this "guilt by association" strategy has identified many antiphage genes, scientists have hypothesized that hundreds more exist.
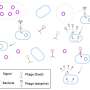
Taking a new tack, Dr. Forsberg, Mr. Rodriguez-Rodriguez, and their colleagues used DNA extracted from bacteria living in human fecal and oral samples, as well as grassland soil samples. After breaking up this DNA into small pieces, each holding about three or four genes, the researchers inserted individual pieces into Escherichia coli, bacteria that is found in human intestines and often used as a lab model.
They then grew these bacteria in petri dishes coated with one of seven types of phages that attack E. coli, looking for microbes that survived phage attack and grew into colonies—an indication that the inserted snippet of DNA held a phage-fighting gene.
This strategy turned up over 200 phage defense systems, including many previously unknown. A closer look showed that several of these genes encoded nucleases, or enzymes that cleave the nucleic acids DNA or RNA. Several more encoded DNA modification-dependent enzymes, generating enzymes that only cleave nucleic acids onto which sugars, proteins, or other molecules are bound—a common strategy some viruses use to avoid bacterial restriction enzymes, another form of phage defense.
Other phage defense genes encoded proteins present on the surfaces of some bacterial species that prevent phages from binding. The researchers also found some genes that appear to cause "abortive infection," in which bacteria become dormant or die after they are infected to protect nearby bacteria from infection.
However, Dr. Forsberg said, the majority of phage defense genes that turned up in the study have unknown functions. He and his colleagues plan to investigate their roles in future research.
Other UTSW researchers who contributed to this study are Vincent Tagliabracci, Ph.D., Associate Professor of Molecular Biology; and James Pfister, B.S., Arabella Martin, B.S., and Luis Mercado-Santiago, B.S., graduate student researchers.
More information: Luis Rodriguez-Rodriguez et al, Metagenomic selections reveal diverse antiphage defenses in human and environmental microbiomes, Cell Host & Microbe (2025).
Journal information: Cell Host & Microbe
Provided by UTÂ Southwestern Medical Center