Biophysical techniques reveal dynamic movements in RNA-degrading molecular machines

Lisa Lock
scientific editor

Robert Egan
associate editor

Researchers at the Regensburg Center for Biochemistry (RCB) and the Regensburg Center for Ultrafast Nanoscopy (RUN) at the University of Regensburg are obtaining unique insights into the structure, dynamics and function of dynamic components of the exosome, an RNA-degrading molecular machine in the cell. The results not only provide biological information on RNA degradation but are also a methodological milestone in structural elucidation of biomolecules.
The work shows that the combination of experimental and computer-assisted biophysical methods facilitates the investigation of dynamics in large molecular machines. Such investigations were not possible before. The interdisciplinary team led by Dr. Jobst Liebau, Dr. Daniela Lazzaretti, Prof. Dr. Till Rudack and Prof. Dr. Remco Sprangers reports on their in Nature Communications.
Proteins are the all-rounders in the cells of every living organism and the basis of all life. Often, several proteins form larger complexes that perform a variety of vital tasks as molecular machines. For example, these complexes assemble vital molecules and disassemble them again when they are not needed anymore. Other complexes transport and sort molecules, or they send and receive messages.
To understand how those protein complexes perform their functions, one must understand what they look like. In recent decades, researchers have elucidated the three-dimensional (3D) structure of a large number of proteins. The University of Regensburg has its own high-resolution cryo-electron microscope available for this purpose. This microscope was used in the current study to elucidate the static structure of a molecular machine.
"These structures are very important, but not sufficient," says Prof. Dr. Sprangers, Professor of Biophysics at the University of Regensburg. "To truly understand the function of proteins, we need to understand how they move and how their structure changes when they perform their function. This is a task that is even more challenging than elucidating the rigid structure."
Sprangers's research aims to do just that. Using nuclear magnetic resonance (NMR) spectroscopy, his research group is investigating how proteins change their structure to perform their function. But how can these changes be visualized? NMR data is often very abstract.
Molecular dynamics (MD) simulations are used to calculate dynamic structural models that visualize the structural changes. However, these models require experimental verification. "The combination of NMR and MD works analogously to a microscope with very high spatial and temporal resolution and provides a kind of movie of the atomic interaction of proteins," explains Prof. Dr. Rudack, Professor of Structural Bioinformatics at the University of Regensburg.
In most cases, however, the NMR method only worked for small proteins. "NMR often reaches its limits with larger protein complexes. We have now achieved a breakthrough that makes it possible to study the giants of the microscopic world of proteins, such as the RNA exosome complex, which plays a crucial role in RNA degradation," explains Dr. Liebau, postdoc in the Sprangers group and first author of the study.
"In addition, we have now been able to study areas of the exosome complex that were previously invisible to all other methods," adds Dr. Lazzaretti, also a postdoc in the Sprangers group.
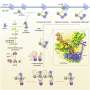
The RNA exosome consists of 10 distinct proteins and degrades RNA. This is an essential task in every cell. "The fact that we can measure the movements of previously invisible regions allows us to analyze the short-lived interactions between RNA and the exosome," explains Dr. Liebau.
Some areas of the protein move extremely fast, performing a movement several billion times per second. Other, mostly larger regions move more slowly: only 30 times per second. It is precisely these slow movements that often appear to be of central importance for the function of protein complexes.
For example, the researchers were able to identify an area in the RNA exosome that moves at roughly the same speed as the exosome degrades RNA. The researchers have not yet been able to prove a direct connection, but without movement, there would be no RNA degradation.
The study thus provides more than a static image. Rather, it provides a kind of movie that offers insights into the dynamic processes of RNA degradation by the exosome complex. "Life is movement," explains Prof. Dr. Rudack, "and the interplay of the NMR method and MD simulations provides deep insights into the dynamic world of proteins."
"The combination of different biophysical methods to elucidate structural dynamics is groundbreaking for future research. We are only just beginning to understand the role that dynamics plays in the function of proteins," adds Prof. Dr. Sprangers.
With their study, the researchers have laid the foundation for transforming the previously static images of the microcosm of the cell into moving ones.
More information: Jobst Liebau et al, 4D structural biology–quantitative dynamics in the eukaryotic RNA exosome complex, Nature Communications (2025).
Journal information: Nature Communications
Provided by University of Regensburg