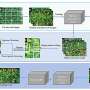
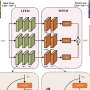
Deep learning tool sets benchmark for accurate rice panicle counting across growth stages

Gaby Clark
scientific editor

Robert Egan
associate editor

By integrating large-kernel convolutional blocks and a novel loss function, LKNet effectively addresses challenges such as overlapping targets, annotation bias, and variability in panicle structure across growth stages. Tested on UAV imagery and multiple crop datasets, the model demonstrates superior performance and robustness, offering a high-throughput solution for precision agriculture and crop.
Rice panicle counting has historically relied on detection-, density-, or location-based methods. However, detection models struggle in crowded canopies, while density-based models are sensitive to background interference. Location-based methods such as P2PNet offer simplicity and interpretability but are limited by receptive field constraints and label inaccuracies.
These challenges are compounded by panicle variations across rice types and growth stages. Addressing these issues, the new LKNet model extends P2PNet's framework with dynamic receptive field adaptation and a more flexible loss function to improve counting robustness and accuracy.
A published in Plant Phenomics by Song Chen's team, Chinese Academy of Agricultural Sciences, supports a range of agricultural applications—from yield prediction to breeding phenotyping—without the need for time-intensive manual annotations.
To evaluate the effectiveness of the proposed LKNet model, the research team conducted a series of comparative and ablation experiments across multiple datasets and counting tasks. The model, designed with large-kernel convolutional blocks (LKconv) and an optimized localization loss function, was first benchmarked against existing methods on both crowd and crop counting datasets.
On the high-density SHTech PartA crowd dataset, LKNet achieved a mean absolute error (MAE) of 48.6 and root mean square error (RMSE) of 77.9, outperforming both P2PNet and the detection-based PSDNN_CHat model. On the PartB dataset, LKNet matched state-of-the-art performance with minimal error. In cross-domain evaluations, LKNet demonstrated superior accuracy in rice panicle counting (RMSE = 1.76, R² = 0.965), particularly excelling over models that performed better on larger targets such as maize tassels.
Testing LKNet on a rice canopy dataset collected at 7 meters showed consistently high performance across panicle types—compact, intermediate, and open—with R² values exceeding 0.98. However, accuracy declined slightly at later growth stages due to increasing occlusion and morphological variation.
Ablation studies confirmed that integrating the LKconv backbone significantly improved both accuracy and model efficiency, reducing RMSE from 2.821 to 0.846 while cutting parameter count by nearly 50%. Moreover, among various kernel designs, the sequential large-kernel module with attention mechanism yielded the highest accuracy (R² = 0.993).
Visualization of class activation maps further illustrated LKNet's enhanced ability to localize panicles, especially in complex scenes. Compared to P2PNet, LKNet exhibited broader focus areas and better background suppression. Overall, the results demonstrate that LKNet not only surpasses current models in precision and efficiency but also adapts well to the diverse spatial and phenotypic complexities inherent in field-based rice canopy counting.
LKNet represents a significant advancement in UAV-based crop monitoring, delivering precise and efficient rice panicle counting across diverse field conditions. Its innovations in model architecture and loss function allow for dynamic adaptation to real-world variability, making it a powerful tool for agricultural phenotyping.
More information: Ziqiu Li et al, LKNet: Enhancing rice canopy panicle counting accuracy with an optimized point-based framework, Plant Phenomics (2025).
Provided by Chinese Academy of Sciences