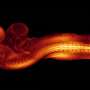
These four images show patterns of gene expression in the neural plates of normal (left) and Sonic Hedgehog mutant (right) embryos. Understanding how these genes are correctly patterned in space is a critical step in understanding how the brain is formed. In this study, the authors examined gene expression in individual cells to understand how these patterns arise. Credit: North Carolina State University
Studying the process of brain formation illuminates just how much of development is a series of tiny miracles. Only a few weeks after a human egg is fertilized, a sheet of cells called the neural plate widens, stretches and rolls up to create a tube. This delicate dance of cells forms what will become our brain and spinal cord, the basis for our thoughts, feelings and actions.
How the cells "know" what they will become is not well understood, but new research by Eric Brooks, a developmental biologist at the NC State College of Veterinary Medicine, and his colleagues offers clues.
By studying how RNA expression patterns change across cells over time in very early mouse embryos, the scientists developed a map of the neural plate and followed it through time as it formed the neural tube. In the process, Brooks and his colleagues showed that they can pinpoint where a cell will move in the early brain based entirely on the RNA that cell is expressing.
Published in eLife, could help scientists around the world as they work to understand brain development—and how to stop that development from going wrong.
All embryos start as a ball of cells. Those cells multiply and differentiate over time into the tissues we recognize as our muscles, bones, brain and organs. The cells that will become the central nervous system—the brain and spinal cord—are some of the first to get into formation.
Brooks has always been curious to understand how the cells know where to go, how they understand what their eventual place and role will be.
"A lot of people are fundamentally interested in where the brain comes from," Brooks says. Scientists know that chemical signals form gradients—concentrations that help guide cells to where they need to be. But the changes happening inside those cells—changes that will make those cells into different types of neurons and support cells, all interacting with one another in highly specific ways—remain a mysterious dance.
"What is it that's really helping to differentiate these cells from one another?" Brooks asks. "Or allow them to do specific things like create the shape of the tissue?"
Working with computational biologists
The complicated choreography offers many opportunities for missteps.
"Defects in the formation of this structure, the cranial neural tube, or the neural tube more broadly, are among the most common developmental defects in humans," Brooks says, affecting one in every 2,000 births.
If the defects are in the part of the tube that forms the eventual spinal cord, the result can be types of spina bifida. But if the tube doesn't close in the cranial region, which would become the brain, "that's not survivable," Brooks says.
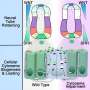
To better understand the paths that cells take and how they achieve their eventual fates, Brooks collaborated with the labs of computational biologist Dana Pe'er and developmental biologist Jennifer Zallen, Howard Hughes Medical Institute researchers at the Memorial Sloan Kettering Cancer Center, to examine RNA expression in the cells of a mouse cranial neural plate.
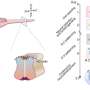
Construction of a single-cell RNA expression atlas during mouse cranial neural tube closure. Credit: eLife (2025). DOI: 10.7554/eLife.102819.3
Inside every cell, DNA offers the same instruction manual. Each cell pulls different information from the manual to carry out its tasks by transcribing the instructions into RNA. By tracking what RNA a cell is expressing, scientists can tell which genes the cell is using to guide its activities.
Brooks and his colleagues carefully isolated cells from mouse embryos between 7.5 and 9 days after fertilization—the time when the neural plate expands and closes to form the neural tube in mice. They used a technique called single-cell RNA sequencing to tease more than 17,000 cells apart and show which RNA was active in each cell.
The scientists were then able to construct a set of maps showing where different cells were expressing specific RNA at different points in time. The result is a beautiful cellular atlas, with colors washing over different areas at specific points in time as the cells divide and change over 1.5 days.
Brooks and his colleagues showed that the RNA expression of each cell could be used to determine where a cell came from—in the forebrain, midbrain or hindbrain. Other patterns of expression showed whether cells came from the middle of the plate or toward the outer edge.
The Sonic Hedgehog gene
The scientists were also interested in a particular gene called Sonic Hedgehog. The expression of this gene sets off a cascade of other genetic signals.
"Sonic Hedgehog itself is expressed in a kind of narrow stripe right in the midline of the tissue," Brooks explains.
Too much Sonic Hedgehog signaling can cause problems with neural tube closure—cutting off the development of early brain structures. With their RNA sequencing atlas, Brooks and his colleagues were able to , showing how overexpression of Sonic Hedgehog affected the neural tube. Mutations in the gene stopped the edges of the neural plate from folding in and forming the tube in the first place.
"One of the really fascinating things about development is it's generally kind of progressive," Brooks says. "You're building on things that have already happened. And so the stages that we highlighted in this atlas are really important."
The RNA map shows how the cells begin to organize themselves and also provides clues as to what their cellular futures will be, what roles they might play and connections they might eventually form.
Brooks hopes other scientists will also use this atlas to ask questions about very early brain development. "This is definitely what we think of as a resource for the community," he says.
Many questions remain about how the cell layers of the early embryo interact. With the help of the atlas, scientists may be able to better understand how an embryo goes from a few layers of simple cells to the vast complexity of the fully formed human brain.
More information: Eric R Brooks et al, A single-cell atlas of spatial and temporal gene expression in the mouse cranial neural plate, eLife (2025).
Journal information: eLife
Provided by North Carolina State University