Scientists map root-released molecules to guide development of hardier bioenergy crops

Lisa Lock
scientific editor

Andrew Zinin
lead editor

Scientists at the Department of Energy's Oak Ridge National Laboratory conducted a meticulous analysis of the compounds released by plant roots into their surrounding environment. The analysis yielded an abundance of data that can guide research aiming to improve the way we grow energy and food crops.
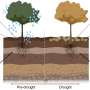
Researchers have long known that the relationships plants form with microbes like bacteria and fungi can make plants more tolerant of poor growing conditions, such as drought or scant nutrients. As plants grow, they release organic molecules into the soil, a process known as rhizodeposition. This organic matter in turn affects how plants and microorganisms interact with other belowground processes.
ORNL scientists developed a new analytical framework based on metabolomics—the study of small molecules—to systematically characterize plant-derived rhizodeposits. The work produced much data about the diversity and relative amounts of compounds in soils, as described in an published in Plant, Cell & Environment.
Information gleaned from the project enhances the understanding of interactions among plants and microbiomes to guide the development of higher-yielding, stress-resistant varieties of crops, and the engineering of microbes that aid in crop resilience. The results enable the development of hardy, productive bioenergy feedstocks for the bioeconomy, strengthening domestic supply chains and energy security.
Untargeted approach expands results
Researchers devised an experiment in which two varieties of poplar trees were grown in controlled conditions, with and without added nutrients. Samples were taken from actively growing and more established root areas at different times, with researchers using an approach called untargeted metabolomics that allowed them to detect and analyze as many molecules as possible, not just a pre-selected few.
The team used high-resolution mass spectrometry to identify and quantify chemical compounds, generating a molecular fingerprint of the samples. Computational methods were then used to group and compare the compounds.
The result was a treasure trove of rhizodeposit compounds, many never before identified. Their composition varied depending on the plant type, nutrient availability, location and time. Researchers drew on ORNL's extensive genomic data on poplar, a key bioenergy crop of interest, to understand how genetics played a major role in shaping those compounds.
"Metabolomics has mostly been limited to targeted analysis, confirming a specific compound or interaction you suspect is in the sample," said project co-lead Paul Abraham of ORNL's Biosciences Division. "But with an untargeted approach, we can capture a much broader range of chemical diversity, revealing unexpected or previously unrecognized compounds that may play critical roles in soil and plant systems."
Next steps: AI-assisted discovery
"This project was made possible by ORNL's ultra-precise mass spectrometry instruments and interdisciplinary environment," Abraham said. "The accuracy and sensitivity of these capabilities are paramount to the success of untargeted metabolomics. Our team of experts in genomic science, plant systems biology and bioanalytical chemistry were essential to designing and executing the study and understanding the implications of the work."
Follow-on research may include the deployment of AI tools to analyze the data, Abraham added. "The chemical space we are measuring is vast, and most of the molecules we detect can't be confirmed using existing reference standards," he said.
"To make sense of that complexity, we'll increasingly depend on machine learning and AI to resolve chemical formulas into predicted structures. That's why one of our key goals is to make our data findable, accessible and reusable for the broader scientific community."
Scientists could also leverage the digital underground root analytics system being installed at ORNL's Advanced Plant Phenotyping Laboratory to enable image-based analysis of root system dynamics, potentially extracting even more features, he added.
More information: Manasa R. Appidi et al, Dynamic Rhizodeposition in the Woody Perennial Populus trichocarpa, Plant, Cell & Environment (2025).
Provided by Oak Ridge National Laboratory