Computational method cuts through the noise to bring clarity to single-cell analysis

Sadie Harley
scientific editor

Robert Egan
associate editor

The world of cells is surprisingly noisy. Each cell carries unique genetic information, but when we try to measure cellular activity, signals can be lost or blurred, and differences between experiments can further obscure the data. These challenges have made it difficult for researchers to capture the true behavior of cells, especially when studying rare cell types or subtle changes that appear in the early stages of disease.
Take single-cell RNA sequencing as an example. It is a powerful technique for studying gene expression at the individual cell level, yet often encounters significant challenges due to two main types of noise: technical noise and batch noise.
Technical noise arises from inherent limitations in measurement processes, such as the "dropout effect," where certain genes are not detected even if they are expressed. Batch noise refers to variations introduced during experiments, like differences in experimental conditions or equipment used, leading to inconsistencies across datasets.
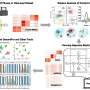
To address these challenges, Associate Professor Yusuke Imoto and his team at Kyoto University's Institute for the Advanced Study of Human Biology (WPI-ASHBi) developed RECODE (resolution of the curse of dimensionality), a high-dimensional statistical method that reduces technical noise in single-cell RNA-sequencing data.
Single-cell data is "high-dimensional," meaning that thousands of genes are measured in each cell. In such high-dimensional spaces, any random noise can overwhelm true biological signals, a problem known as the "curse of dimensionality." Traditional statistical methods struggle to identify meaningful patterns under these conditions.
RECODE overcomes this problem by applying advanced statistical methods to reveal expression patterns for individual genes close to their expected values. This approach has been shown to outperform other methods, providing clear gene activation profiles without relying on complex parameters or machine learning techniques.
Building on RECODE, Prof. Imoto has now introduced iRECODE (Integrative RECODE), an enhanced version that simultaneously reduces both technical and batch noise with high accuracy and low computational cost.
This improvement enables more comprehensive noise reduction, making it easier to detect rare cell types and subtle biological changes critical for understanding complex processes.
iRECODE works across multiple types of single-cell datasets, including RNA sequencing, spatial transcriptomics, and scHi-C, revealing cellular patterns that were previously hidden. These findings were online in Cell Reports Methods.
Key findings
When applied to single-cell RNA sequencing data, iRECODE refined the gene expression distributions and resolved sparsity (where many data entries are 0, caused by technical noise). Also, iRECODE effectively reduces the batch noise, achieving better cell-type mixing across batches while preserving each cell type's unique identity.
The method is approximately 10 times more efficient than the combination of technical noise reduction and batch correction methods, and it works across multiple technologies including Drop-seq, Smart-Seq, and multiple 10x Genomics protocols.
RECODE's capabilities extend beyond single-cell RNA sequencing, as it can help reduce technical noise in other single-cell datasets that rely on random molecular sampling. For example, scHi-C data measure how different parts of a chromosome physically interact in individual cells.
However, scHi-C data can be very sparse, making it difficult to identify meaningful cell types or chromosomal contacts. Applying RECODE greatly reduces this sparsity and uncovers real interactions that better reflect differences between cells. Furthermore, a combination of RECODE and an existing machine learning-based method accelerates the accuracy of cell clustering.
Another example of RECODE's application is spatial transcriptomics, which allows us to study how different cells behave and interact within tissues, though technical noise can blur important patterns. Across different platforms, species, tissue types, and genes, RECODE consistently clarified signals and reduced sparsity, demonstrating its broad applicability in spatial transcriptomics.
iRECODE allows researchers to "listen" to the true voices of individual cells, making it possible to study cellular behavior with unprecedented clarity. Its ability to handle multiple data types and large-scale datasets positions it as a key tool for future single-cell analysis and a potential standard preprocessing step for single-cell studies.
Researchers anticipate that iRECODE will help uncover previously hidden biological patterns, such as rare cell populations and subtle changes associated with aging or early disease stages.
"Single-cell data captures countless cellular 'whispers,' but hearing those whispers through the noise is extremely difficult," commented Prof. Imoto.
"iRECODE, an evolution of our RECODE method, lifts those voices to the surface. Through this method, I believe the hidden stories of cells, stories we could never hear before, will steadily come to light."
More information: Comprehensive noise reduction in single-cell data with the RECODE platform, Cell Reports Methods (2025). .
Journal information: Cell Reports Methods
Provided by Kyoto University