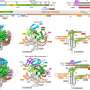
Discovery of TranC systems reveals the molecular mechanism of CRISPR origin. Credit: IGDB
CRISPR-Cas systems are adaptive immune systems found in prokaryotes that defend against invading nucleic acids through CRISPR RNA-guided cleavage. Type V CRISPR-Cas (Cas12) systems, in particular, serve as one of today's most powerful tools for genome editing, especially in basic research, medicine, and agriculture.
Researchers led by Prof. Gao Caixia from the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS), together with Assoc. Prof. Liu Junjie from Tsinghua University and Prof. Zhang Yong from the Institute of Zoology of CAS, have now uncovered the molecular innovation that led to the origin of type V CRISPR-Cas immune systems.
Their findings, published on September 29 in Cell, show that the functional splitting of transposon-derived RNAs was the critical innovation driving the emergence of type V CRISPR-Cas immunity.
Previous studies showed that the ancestral proteins of type V Cas12 effectors are TnpB nucleases encoded by IS200/605 transposons. However, the molecular mechanisms bridging transposon activity and CRISPR immunity had remained unclear.
To address the origin of type V CRISPR-Cas systems, the researchers developed a unified mining strategy that combined catalytic motifs, structural domains, and sequence similarity shared between the TnpB and Cas12 nucleases.
By searching prokaryotic genomes and metagenomic databases, they identified 146 TnpB-like CRISPR-associated proteins. Through phylogenetic analyses, AlphaFold-based structural predictions, and functional element comparisons, the researchers ultimately identified six intermediate clades, collectively called TranCs, which form sister groups to specific TnpB lineages. Notably, clades 3, 11, 12, 13, and 14 originate from IS605, whereas the previously reported clade 8 (Cas12n) arises from IS607, representing key evolutionary intermediates between TnpB and Cas12.
Functional assays revealed a dual-guide RNA mechanism unique to TranCs. The researchers found that five TranC systems not only utilized their intrinsic CRISPR RNAs (tracrRNA-crRNA hybrids) for DNA targeting but also retained the ancestral capacity to utilize transposon-derived reRNAs (also called ωRNAs) to direct DNA cleavage. This dual-guidance capacity provides a functional signature showing TranCs as evolutionary intermediates.
Cryo-EM analysis of the LaTranC-sgRNA-DNA complex also revealed a striking resemblance to the ISDra2 TnpB-reRNA-DNA complex, with one crucial difference: The single reRNA had undergone functional splitting into two components, tracrRNA and crRNA. The reRNA and CRISPR RNA covariance analyses and AlphaFold protein model comparison extended this observation across three clades from IS605 and IS607, establishing RNA splitting as a shared hallmark of Cas12 emergence.
Importantly, engineering experiments confirmed that artificially splitting TnpB's reRNA was sufficient to convert TnpB into a CRISPR-like system capable of using CRISPR arrays as a source of guide RNAs. These results demonstrate that RNA-level innovation, rather than major protein structural changes, was the primary molecular event driving the origin of type V CRISPR-Cas systems.
This study is significant not only for clarifying the molecular mechanism behind the evolution of type V CRISPR systems, but also for identifying a set of compact nucleases with flexible guide RNAs, offering design principles to develop CRISPR tools that are smaller, more versatile, and easier to control.
More information: Functional RNA splitting drove the evolutionary emergence of Type V CRISPR-Cas systems from transposons, Cell (2025).
Journal information: Cell
Provided by Chinese Academy of Sciences