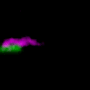
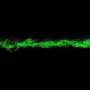
Bacteria confined to droplets form complex but predictable patterns based on oxygen levels

Sadie Harley
scientific editor

Robert Egan
associate editor

Even in an environment as seemingly simple as a drop of water, bacteria can organize themselves into complex patterns or arrangements. Through experiments, theory, and computational modeling, scientists from Caltech and Princeton University have found that the way these cells arrange themselves is shaped by oxygen levels and is predictable and controllable, offering insights that could lead to new ways to target infections.
"Imagine a drop of a fluid with bacteria uniformly swimming around within it, like a little bacterial dance floor," says Sujit Datta, professor of chemical engineering, bioengineering, and biophysics at Caltech.
The bacteria are confined naturally by the perimeter of the droplet—a situation that could apply to bacterial cells in any liquid film, such as those found in the mucus lining of the lungs, or in tiny drops of moisture in the soil. The cells need oxygen to keep moving around the dance floor.
"If there are enough of those bacteria on the dance floor, the ones near the perimeter are going to consume the oxygen first," Datta says.
"And if the decrease in oxygen is large enough—say, if there are too many bacteria or the droplet is really large—the ones in the middle just don't get enough oxygen, and they essentially fall asleep."
The cells at the center of the droplet become dormant, forming a dense core surrounded by a perimeter of cells that keep "dancing" because they still have access to oxygen.
Datta and his colleagues describe their observations of these arrangements, and why they form, in a in the journal PNAS. The lead author of the paper, "Spatial self-organization of confined bacterial suspensions," is Babak Vajdi Hokmabad, who worked as a postdoctoral scholar in Datta's group.

The scientists used a confocal fluorescence microscope to observe bacteria in trapped droplets with high spatial resolution and in three dimensions. Surprisingly, Datta says, if the droplets are big enough and the concentration of bacteria high enough, the complex arrangements formed by the dormant and still-active cells can be seen with the naked eye.
Guided by the experiments, Datta and his team worked out a mathematical model that draws on ideas from biology, chemistry, and physics to account for the processes behind what they were seeing. Between the models and additional careful experiments, they were then able to predict when they could expect to see bacteria in a particular droplet form these types of patterns and observe how those arrangements might change over time.
"In an interesting twist, it turns out the active cells move toward regions with even more oxygen, near the edge of a droplet. And in doing so, they can actually change the rate at which oxygen can get in," Datta explains. "In some cases, by doing that, they promote further influx of oxygen, and that actually causes the dormant cells to eventually wake up."
Datta says the work is exciting at a fundamental level because biological physicists are interested in how seemingly simple organisms can create complex structures as collectives. But there is also a range of areas where the findings could be applied. For example, he says, both aerobic and anaerobic bacteria are found in the lining of the lungs.
The anaerobic bacteria can be particularly dangerous because they tend to be more metabolically dormant, a state that can allow them to more effectively evade antimicrobial agents.
"The question is: Can we come up with quantitative guidelines for how to improve the efficacy of antimicrobial treatments that often miss these because they rely on metabolic activity and oxygen to be effective?" Datta says. "We've just scratched the tip of the iceberg, but that's one context in which I think the implications of our work are useful."
More information: Babak Vajdi Hokmabad et al, Spatial self-organization of confined bacterial suspensions, Proceedings of the National Academy of Sciences (2025).
Journal information: Proceedings of the National Academy of Sciences
Provided by California Institute of Technology