A new system can dial expression of synthetic genes up or down

Lisa Lock
scientific editor

Robert Egan
associate editor

For decades, synthetic biologists have been developing gene circuits that can be transferred into cells for applications such as reprogramming a stem cell into a neuron or generating a protein that could help treat a disease such as fragile X syndrome.
These gene circuits are typically delivered into cells by carriers such as nonpathogenic viruses. However, it has been difficult to ensure that these cells end up producing the correct amount of the protein encoded by the synthetic gene.
To overcome that obstacle, MIT engineers have designed a new control mechanism that allows them to establish a desired protein level, or set point, for any gene circuit. This approach also allows them to edit the set point after the circuit is delivered.
"This is a really stable and multifunctional tool. The tool is very modular, so there are a lot of transgenes you could control with this system," says Katie Galloway, an assistant professor in Chemical Engineering at MIT and the senior author of the new study.
Using this strategy, the researchers showed that they could induce cells to generate consistent levels of target proteins. In one application that they demonstrated, they converted mouse embryonic fibroblasts to motor neurons by delivering high levels of a gene that promotes that conversion.
MIT graduate student Sneha Kabaria is the lead author of the , which is published today in Nature Biotechnology. Other authors include Yunbeen Bae '24; MIT graduate students Mary Ehmann, Brittany Lende-Dorn, Emma Peterman, and Kasey Love; Adam Beitz Ph.D. '25; and former MIT postdoc Deon Ploessl.
Dialing up gene expression
Synthetic gene circuits are engineered to include not only the gene of interest, but also a promoter region. At this site, transcription factors and other regulators can bind, turning on the expression of the synthetic gene.
However, it's not always possible to get all of the cells in a population to express the desired gene at a uniform level. One reason for that is that some cells may take up just one copy of the circuit, while others receive many more. Additionally, cells have natural variation in how much protein they produce.
That has made reprogramming cells challenging because it's difficult to ensure that every cell in a population of skin cells, for example, will produce enough of the necessary transcription factors to successfully transition into a new cell identity, such as a neuron or induced pluripotent stem cell.
In the new paper, the researchers devised a way to control gene expression levels by changing the distance between the synthetic gene and its promoter. They found that when there was a longer DNA "spacer" between the promoter region and the gene, the gene would be expressed at a lower level. That extra distance, they showed, makes it less likely that transcription factors bound to the promoter will effectively turn on gene transcription.
Then, to create set points that could be edited, the researchers incorporated sites within the spacer that can be excised by an enzyme called Cre recombinase. As parts of the spacer are cut out, it helps bring the transcription factors closer to the gene of interest, which turns up gene expression.
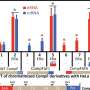
The researchers showed they could create spacers with multiple excision points, each targeted by different recombinases. This allowed them to create a system called DIAL, that they could use to establish "high," "med," "low" and "off" set points for gene expression.
After the DNA segment carrying the gene and its promoter is delivered into cells, recombinases can be added to the cells, allowing the set point to be edited at any time.
The researchers demonstrated their system in mouse and human cells by delivering the gene for different fluorescent proteins and functional genes, and showed that they could get uniform expression across a population of cells at the target level.
"We achieved uniform and stable control. This is very exciting for us because lack of uniform, stable control has been one of the things that's been limiting our ability to build reliable systems in synthetic biology. When there are too many variables that affect your system, and then you add in normal biological variation, it's very hard to build stable systems," Galloway says.
Reprogramming cells
To demonstrate potential applications of the DIAL system, the researchers then used it to deliver different levels of the gene HRasG12V to mouse embryonic fibroblasts. This HRas variant has previously been shown to increase the rate of conversion of fibroblasts to neurons. The MIT team found that in cells that received a higher dose of the gene, a larger percentage of them were able to successfully transform into neurons.
Using this system, researchers now hope to perform more systematic studies of different transcription factors that can induce cells to transition to different cell types. Such studies could reveal how different levels of those factors affect the success rate, and whether changing the transcription factor levels might alter the cell type that is generated.
In ongoing work, the researchers have shown that DIAL can be combined with a system they previously developed, known as , that uses a feedforward loop to help prevent cells from overexpressing a therapeutic gene.
Using these systems together, it could be possible to tailor gene therapies to produce specific, consistent protein levels in the target cells of individual patients, the researchers say.
"This is something we're excited about because both DIAL and ComMAND are highly modular, so you could not only have a well-controlled gene therapy that's somewhat general for a population, but you could, in theory, tailor it for any given person or any given cell type," Galloway says.
More information: Sneha R. Kabaria et al, Programmable promoter editing for precise control of transgene expression, Nature Biotechnology (2025).
Journal information: Nature Biotechnology
Provided by Massachusetts Institute of Technology
This story is republished courtesy of MIT News (), a popular site that covers news about MIT research, innovation and teaching.