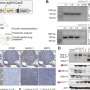
Yeast cells whose cell walls have been stained blue. A protein is marked green as an example. The amount of this or other proteins presentdiffers from cell to cell. The new study has now shown the genetic basis for such variation. Credit: Charité | Jakob Vowinckel
Every organism's genome contains mutations that often have unknown biological effects. In partnership with Stanford University, researchers at Charité—Universitätsmedizin Berlin have now discovered a way to predict the effects of numerous mutations in yeast.
Key to this discovery was a detailed analysis of the proteome—the collection of all proteins inside a cell. The research team believes this new method to be a valuable tool for better understanding molecular mechanisms, for example, in the context of microorganisms' increasing resistance to medicinal agents. The study has been in the Science journal.
Microorganisms are masters of adaptation. Even the smallest of genetic variations can help them adapt to changing, and sometimes hostile, conditions in their habitat. That includes, for example, developing resistance to medicinal agents.
"To be able to better assess the risk of a pathogen becoming resistant, or to develop new and improved agents, we need to learn to better understand the association between different gene variants and their resulting biological mutations," says Prof. Markus Ralser, Director of the Institute of Biochemistry at Charité and one of the two study leads.
"Since genome sequencing has advanced so rapidly, we can identify genetic differences very well nowadays. However, we often do not know their impact on a microbe's growth or resistance, for example, or under which conditions they are significant."
A glimpse into the molecular blackbox
To understand the effects of different gene variants, it helps to take a look at the proteome. The proteome works like a kind of gear train that controls and conducts cellular processes, keeping everything running. The various proteins interlock, almost like cogs, and influence each other.
"For example, a specific variant in a gene can mean that a protein is no longer produced, or is produced in a different form or quantity. And that can actually change quite a lot in the cell's inner workings," says Dr. Johannes Hartl of the Berlin Institute of Health at Charité (BIH) and one of the lead authors of the study. "As its natural genetic variation causes it to be so variable, the proteome is still largely a molecular blackbox. Our study was able to show that it was both possible and necessary to shine more light into this darkness."
The researchers used two naturally occurring strains of yeast cells for their investigation. Yeasts are single-celled microorganisms belonging to the fungi kingdom. One of the yeast strains came from a Californian vineyard, while the other was isolated from an immunosuppressed patient in Italy. The researchers crossbred the two strains over multiple generations.
"This created almost a thousand new yeast strains, in which the parents' genetic features were thoroughly mixed," explains Hartl.
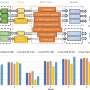
The crossbreeding experiments and subsequent genetic analysis of the yeast strains were carried out in the laboratory at Stanford. The Charité team led by Ralser analyzed the proteome of the different strains using a high-throughput screening process and mass spectrometry. This allowed them to clearly identify different proteins and precisely quantify their respective amounts in the cell.
Proteome reveals molecular basis
Together, the researchers worked through the vast treasure trove of data. The goal: to find clear associations between the individual gene variants and the resulting changes in the proteome.
"To achieve this, we compared the genome and proteome data and created a kind of map that shows the effect of thousands of genetic variants on the amount of thousands of proteins in the cell," Hartl explains.
"And to check whether the associations we found actually stemmed from this particular gene variant, and not from other processes within the cell, for example, we used the CRISPR/Cas 'genetic scissors' to insert the gene variant into the original parent strain of the yeast, which did not previously contain this gene variant. We then looked to see if the corresponding changes in the proteome could also be found here."
The researchers went one step further with some gene variants and the associated changes in the proteome, and they examined their specific effects. For example, they investigated whether the yeast cells could survive under the effects of an antimycotic agent—also known as an antifungal drug.
"The antmycotic agent binds to and inhibits an enzyme that is necessary for the biosynthesis of an essential part of the yeast membrane. This stops the cell from continuing to grow—provided that the agent blocks enough of the present enzymes," says Hartl.
"In our genome-to-proteome map, however, we were able to see that certain gene variants contained raised levels of this enzyme. The experiment showed that yeast cells with this gene variant became more resistant to the antimycotic agent."
Small genetic mutations can have a significant impact
The study also showed that many genetic changes—even those that seem "unremarkable" at first glance—can have far-reaching consequences. The researchers observed that genetic variants affecting hundreds of proteins in the cell had no apparent effect under standard conditions. However, when those conditions were altered, such as through drug treatment or changes in nutrient supply, those variants had a significant impact on cell growth.
"The genome-to-proteome map is an outstanding tool for revealing associations in molecular biology and understanding the impact of mutations and genetic differences," Ralser emphasizes.
"As a result, it is now much easier for us to figure out many of the proteins' functions and interactions, which enables us to better predict how they may potentially develop resistances to agents and adapt to new environments, like humans as host organisms."
In subsequent studies, the researchers therefore wish to extend this approach to fungal pathogens that cause particularly severe infections in humans.
More information: Christopher M. Jakobson et al, A genome-to-proteome map reveals how natural variants drive proteome diversity and shape fitness, Science (2025).
Journal information: Science
Provided by Charité - Universitätsmedizin Berlin