Non-seasonal flu vaccine slides closer to reality
Justin Jackson
сontributing writer

Sadie Harley
scientific editor

Robert Egan
associate editor

University of Pennsylvania researchers have discovered a right-handed, antiparallel double-helix inside influenza ribonucleoprotein complexes that slides strand-against-strand as the viral polymerase copies RNA, offering a conserved target for future antivirals.
Seasonal flu causes 290,000–650,000 deaths annually. Vaccines and most antivirals target viral surface proteins, which mutate rapidly each season, so protection wanes and effectiveness drops. Core viral machinery, nucleoprotein (NP) and RNA polymerase, remains structurally uncharacterized, blocking broad-spectrum drug design.
In the study, "Molecular basis of influenza ribonucleoprotein complex assembly and processive RNA synthesis," in Science, researchers integrated cryo-electron microscopy single-particle analysis and cryo-electron tomography to reveal how NP chains and polymerase coordinate continuous RNA production.
Analysis of 17,414 digital micrographs identified 516,476 particles of a reconstituted influenza D nonstructural segment and 5,738 native influenza A filaments, producing a 5.1 Å reconstruction of four NP subunits and sub-nanometer maps for six additional conformations.
Methods involved assembling the shortest viral segment inside HEK293T kidney cells, purifying complete complexes by tandem affinity and glycerol gradients, then imaging on a 300 kV Titan Krios with graphene-oxide support to protect fragile polymerase-NP contacts.
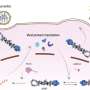
NP organizes into a face-to-face duplex, with RNA threaded through the minor groove and a flexible tail-loop securing adjacent protomers. Polymerase binds the helix exterior, engaging one strand as the two strands slide past each other in an energetically light helical strand-sliding model that preserves overall architecture.
Virtual screening of 30 million compounds against the conserved tail-loop interface identified three candidates: compounds 1, 5 and 23. Compounds 1 and 5 blocked H1N1 replication in MDCK cells without cytotoxicity below 100 µM. Compound 23 showed moderate antiviral activity at low concentrations but became cytotoxic above 50 µM.
Researchers conclude that strand-sliding preserves genome organization and may help the virus evade immune sensors.
Blocking the tail-loop contact could stop that motion, supplying a fresh route towards pan-influenza medicines. Shifting antiviral strategies away from drift-prone surface proteins toward the virus's conserved core machinery could finally yield broad-spectrum flu treatments.
Written for you by our author , edited by , and fact-checked and reviewed by —this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a (especially monthly). You'll get an ad-free account as a thank-you.
More information: Ruchao Peng et al, Molecular basis of influenza ribonucleoprotein complex assembly and processive RNA synthesis, Science (2025). .
Journal information: Science
© 2025 Science X Network